Discover CITRUS: Automatically Identify Predictive Biomarkers
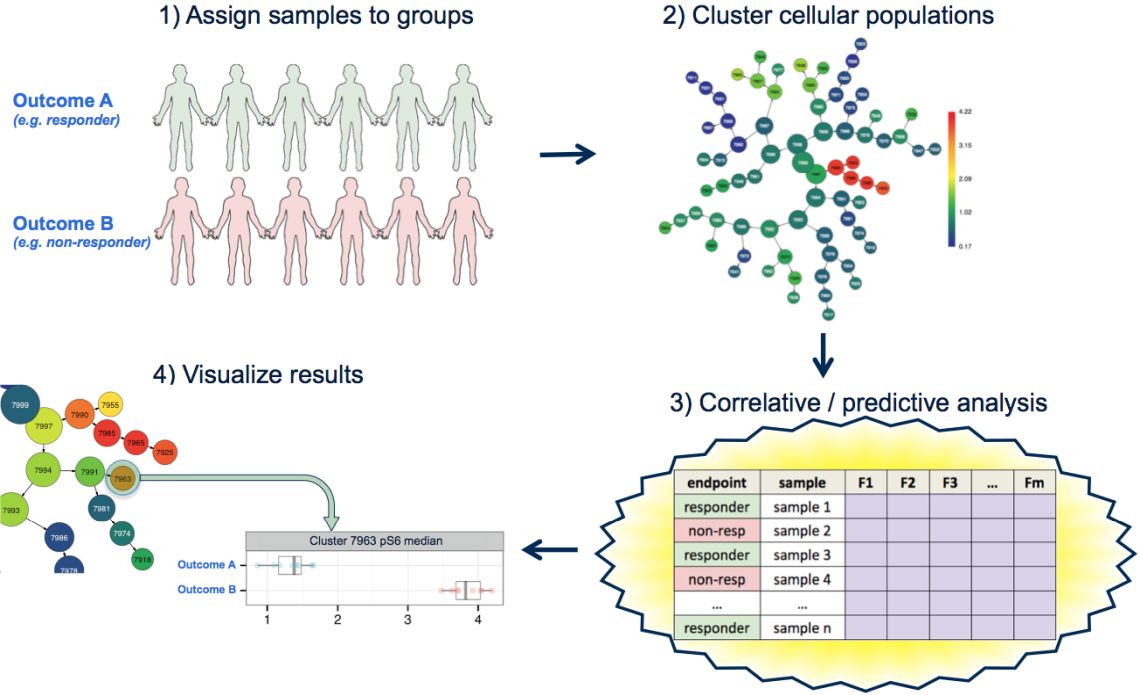
CITRUS (cluster identification, characterization, and regression) is an algorithm that is used for the identification of differential single cell biomarkers.1 It is a black box algorithm that takes a scientific question and returns a set of cell-type specific biomarkers to help answer that question. Importantly, CITRUS does this without relying on prior knowledge or subjective gating to define cell types, and the biomarkers are identified in a statistically sound manner so researchers can be confident that they are following up on true positive results.2
CITRUS is set up to answer two types of questions: ‘How are my groups different?’ or ‘How can I best predict the differences between my groups?’
CITRUS can be used to compare any groups where differences between them are expected to be driven by differences in the abundance of various cell types, the activation or inhibition of signaling or the presence or absence of markers in these cell types. CITRUS will tell the user which of these “features” are significantly different between the defined groups.
CITRUS uses an unsupervised and supervised Machine Learning pipeline to automatically find biomarkers that are correlated with an outcome group or find a set of biomarkers that best predicts that outcome group. The results of a CITRUS run are clusters and associated features that differentiate the observed endpoint of the samples. Watch the video for a tutorial on how to run CITRUS on the Cytobank platform and how to interpret the results.
For more details and instructions also visit our Cytobank Support pages.
References
- Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci USA. 2014;111(26):e2770-e2777. doi:10.1073/pnas.1408792111
- Polikowsky HG, Drake KA. Supervised Machine Learning with CITRUS for Single Cell Biomarker Discovery. In: McGuire HM, Ashhurst TM, eds. Mass Cytometry. Vol 1989. New York, NY: Springer New York; 2019:309-332. doi:10.1007/978-1-4939-9454-0_20