Avoid the Pitfalls When Automating Cell Viability Counting for Biopharmaceutical Quality Control
Abstract
Whether cells are being used to manufacture a therapeutic drug product or drug efficacy testing in pharmacology, measuring the cell viability and concentration accurately and with good reproducibility is crucial. Automated cell counters will count cells according to the technology they use and their individual configuration and this could produce results that vary widely compared both to alternative automated systems and to the common manual technique of using a microscope and haemocytometer. As an example, some people will only include cells as ‘viable’ when calculating cell concentration if the cells are larger than a certain diameter and ignore all the smaller cells, even if the smaller cells’ other characteristics indicate that they are viable. Later in this paper we will show one example of how identifying viable cells by minimum diameter alone can introduce variability of circa 33% in the reported cell concentration. This paper describes how to avoid these common pitfalls when a user is adopting a new automated method of cell counting.
To watch a webinar on this topic, please scroll to the bottom of this page.
Introduction
For therapeutic protein production, getting the concentration of cells correct in the bioreactor is crucial, both to ensure maximum yield and to avoid overcrowding leading to programmed cell death (apoptosis). For drug efficacy testing in pharmacology, it is crucial to get the concentration of cells in every test sample consistent so that the results of the efficacy testing of the drug remain unaffected by potential variability of cell viability and concentration in the samples. In both cases, there is no ‘correct’ way to identify the cells. Users will have optimised their processes based on their own cell counting technique. Some people only count cells as ‘viable’ if they are larger than a certain diameter and ignore all the smaller cells even if they appear to be viable. Later in this paper we will show how identifying viable cells by minimum diameter alone can introduce variability of circa 33% in the reported cell concentration. As another example, some people will count a clump of cells as representing just one single cell, whereas others will try to count the individual cells in the clump.
When moving to an automated cell counting method, it is important to consider how the automated system differentiates viable cells from non-viable cells and cell debris and what impact this may have on your process.
What is the 'correct' answer?
There is no ‘correct answer’ when counting cells. Most people follow the guidance given in EP2.7.291 and USP<1046>2, both of which describe viable cell counting using the dye exclusion method and suggest that Trypan Blue is a suitable dye. Probably the most common technique is the manual method using the haemocytometer slide and a microscope. Variation in results from user to user can be very high with the manual haemocytometer method3 making an automated method appealing. However it is important to take into account how the automated method identifies and counts cells to understand the impact to the process if the results are different between the manual method and the automated method.
Sources of variability in the manual haemocytometer method
There are several steps involved in the manual haemocytometer method. Each can lead to variability and needs to be taken into account when moving to an automated method:
- Accurate pipetting
- Aspiration cycles
- Dye quantity
- Dye mixing cycles
- Size of cells
- Clumps
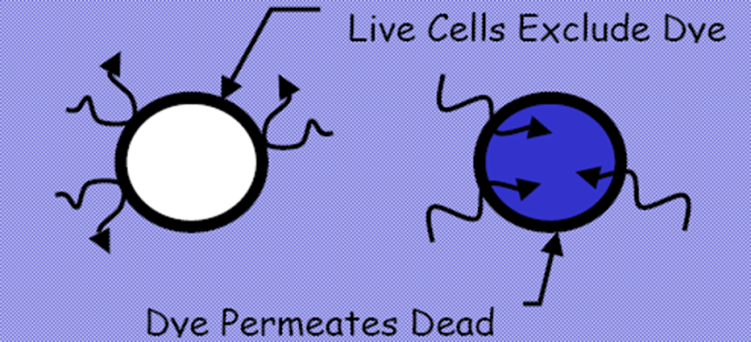
Figure 1. Dye exclusion method from EP 2.7.29 and USP <1046>
Accurate pipetting is crucial. The haemocytometer slide volume is typically 1nL. To report viable cell concentration/ mL, the cell count viewed through the microscope needs to be multiplied by a factor of 100,000 to convert nL to mL, so any small errors in the pipetting volume, and hence cells dispensed onto the slide, will be magnified greatly when the concentration/mL is calculated.
Aspiration can be useful to re-suspend cells and break up clumps. However, techniques vary between users and re-suspension and breaking of clumps will therefore vary. In addition, some cells are particularly delicate and excessive aspiration may actually rupture viable cells’ membranes allowing the dye to enter, resulting in under-counting.
Inaccurate dye addition will impact on the accuracy of the final cell concentration calculation in just the same way that variations in volume through inaccurate pipetting of the sample does as the dye acts effectively as a sample diluent. At the same time, variation in the concentration of the dye may impact on the effective penetration of the dye into the dead and dying cells.
In the same way, variety in the number of dye mixing cycles can have an effect on the penetration of the dye into the dead and dying cells simply because it takes time for the dye to penetrate dead cells and variations in mixing cycles means the cells are exposed to the dye for different amounts of time.
In instances where the health of the viable cells has a large impact on either the pharmacology experiments or on the yield of therapeutic proteins, many users will only count cells as viable when they are larger than a certain size. So dye exclusion is commonly not the only criteria when defining cell viability.
Finally, some users attempt to identify and count all of the individual cells in a clump of cells, whereas others may count the clump of cells as if they were just one cell. The resultant small variations in the way the cells are counted may have a large impact when the concentration/mL is calculated because of the large multiplication factor between nL and mL. The effect of this gets particularly noticeable as the cell concentration/mL increases in the bioreactor and cells tend to clump together more.
Avoiding the common pitfalls
As can be seen, small variations in the way the cells are counted using the manual haemocytometer method can lead to large variations in the resultant calculated viable cell concentration/mL. For this reason alone, many people involved in cell counting have switched to automated cell counting in the search for more reproducible results and, in turn, better controlled pharmacology experiments or manufacturing production yield.
However, there are some common pitfalls: the automated counter must be capable of being adjusted to identify the number of viable cells in the same way as the user would when using a haemocytometer and microscope. The same applies if it is being used to replace some other automated counting system, i.e. it must be capable of being adjusted to identify the number of viable cells in the same way as the other automated counting system it is replacing. If the new automated cell counter is not flexible enough in its counting method, then it will produce a different cell concentration result to the previous method on the same sample of cells, potentially affecting pharmacology experiment results or production yield.
In Figure 2 below, the automated cell counter uses imaging and a series of user-adjustable parameters to decide which cells are viable and which cells are not. The counter allows the user to review each image taken (in this case the analyser is set to take 50 images and average the results). The cell counter identifies for the user which cells it has identified as viable by circling them in green. For non-viable cells it circles them in red.
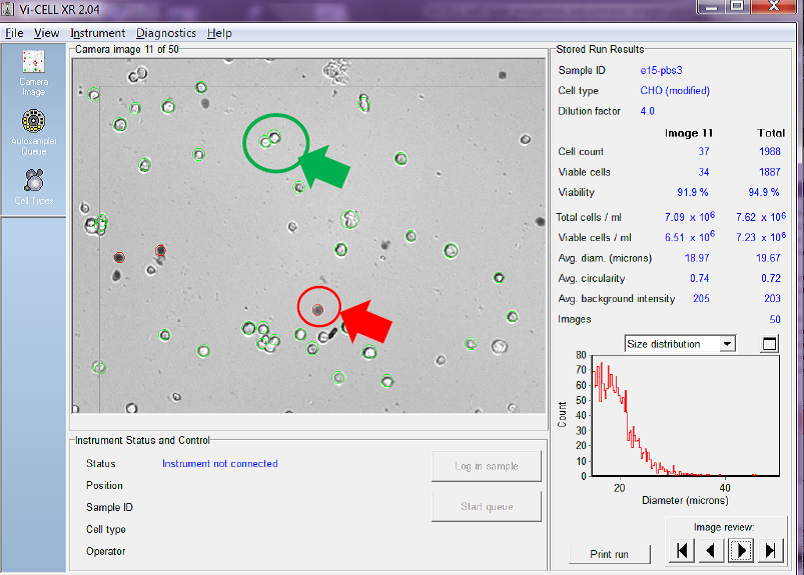
Figure 2. Viable cells are circled in green and non-viable in red
You can also see on the right-hand side of the screen-shot that the counter also calculates the total cell count/ mL, and total viable cell count/mL for both the image being reviewed and as an average of all of the images taken.
Ideally the automated counter should have a wide range of adjustment for the following cell attributes:
a. Cell minimum diameter
b. Cell maximum diameter
c. Cell brightness
d. Cell sharpness
e. Viable cell spot area/size
f. Cell circularity
g. Cell decluster degree
Settings a) and b) allow the user to tell the counter what size of cells they would normally count as being viable.
In the Figure 3 below, the cell images have been analysed using a minimum cell diameter for viable cell identification of 6 micron diameter.
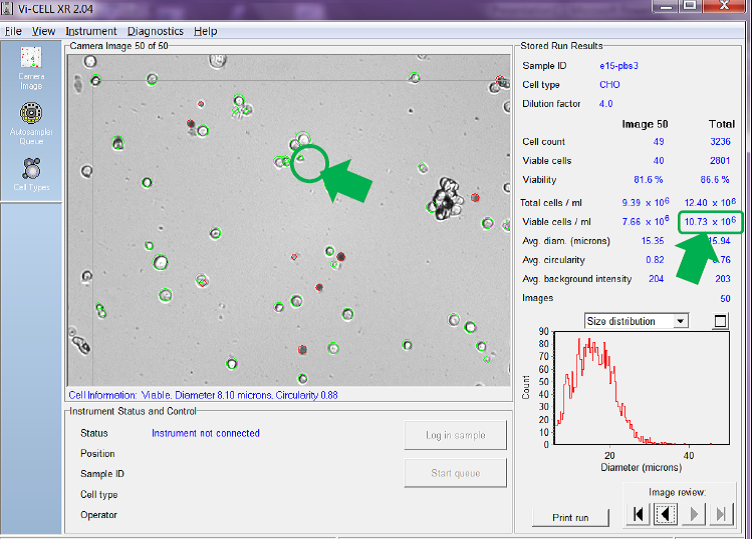
Figure 3. Minimum viable cell diameter set to 6 microns
You can see in Figure 3 that, using the minimum diameter of 6 microns, the counter counts small cells like the one circled which has a diameter of just 8.10 microns. Using this setting the counter has identified an average across the 50 images of 10.73 x 106 viable cells/mL.
If we adjust the minimum diameter to 15 microns so that the counter only counts larger cells as viable and ask the counter to re-analyses the same set of recorded images and report the new calculated cell concentration per mL we get a very different answer (see Fig. 4 below).
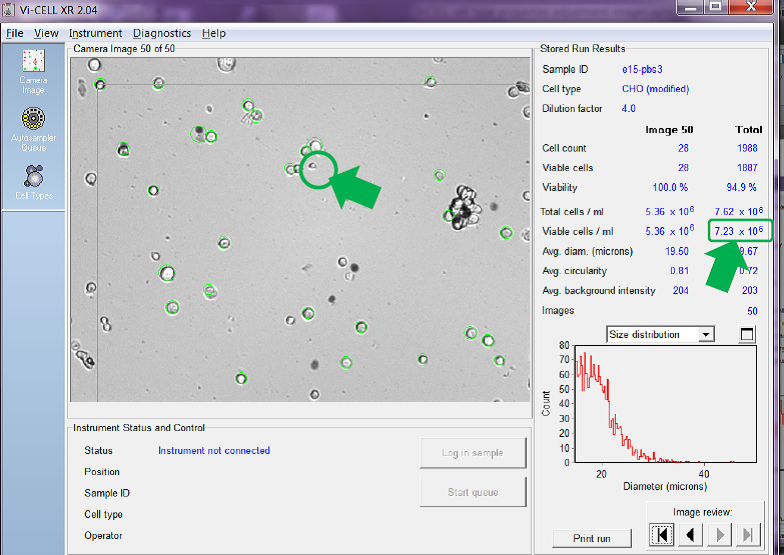
Figure 4. Same images re-analysed with minimum viable cell diameter set
Comparing Fig. 4 to Figure 3 you can see that the counter has no-longer identified the smaller cell in image number 50 as viable (it is not circled in green). The total number of cells in the same set of images counted as viable has been reduced, hence reducing the average calculated viable cell concentration for all 50 images from the 10.73 x 106 viable cells/mL reported in Figure 3 to just 7.23 x 106 viable cells/mL in Figure 4, a difference of 3.5 x 106 viable cells/mL, or a reduction of almost 33%.
Settings c), d) and e) are used to tell the counter how to identify a viable cell, e.g. should a slightly darker cell be counted as viable, or not, etc.
Cell circularity, option f), helps the counter discriminate between any cell debris present and healthy viable cells, but this should be used with caution where viable cells of the cell type used is naturally not spherical in shape.
Finally, setting g) allows the user to instruct the counter whether to count individual cells in a clump, or cluster, as counting to the total cell counts/mL or whether the clump should be counted simply as if it were just one cell.
In Figure 5 below, the counter is set to try to count each and every cell in a clump as an individual cell. You can see that, with this setting, it reports a viable cell concentration of 10.53 x 106 cells/mL.
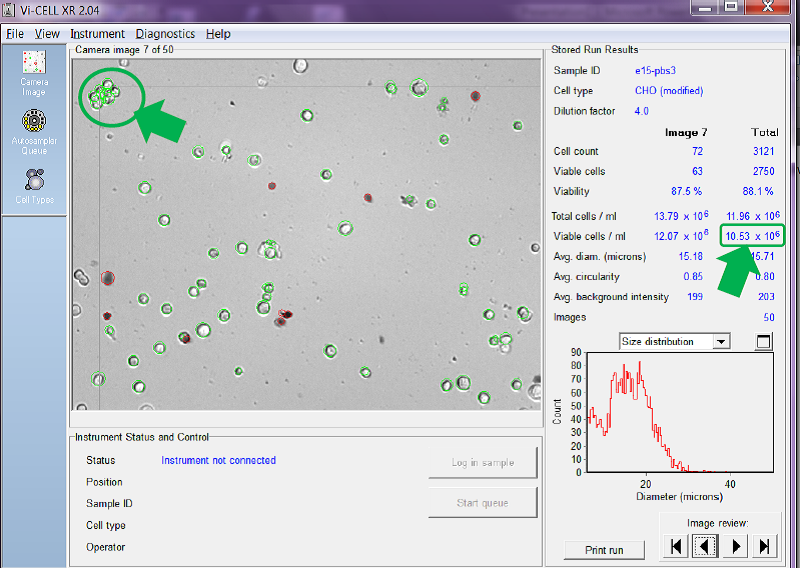
Figure 5. Counter set to count individual cells in clumps to 15 microns
You can see in the clump of cells arrowed in the top left of the image each individual cell has been circled in green and added to the overall viable cell count.
If we adjust the counter to count a clump of viable cells as if it were just one single cell and then ask the counter to re-analyse the same set of images, the answer is very different. See Figure 6 below.
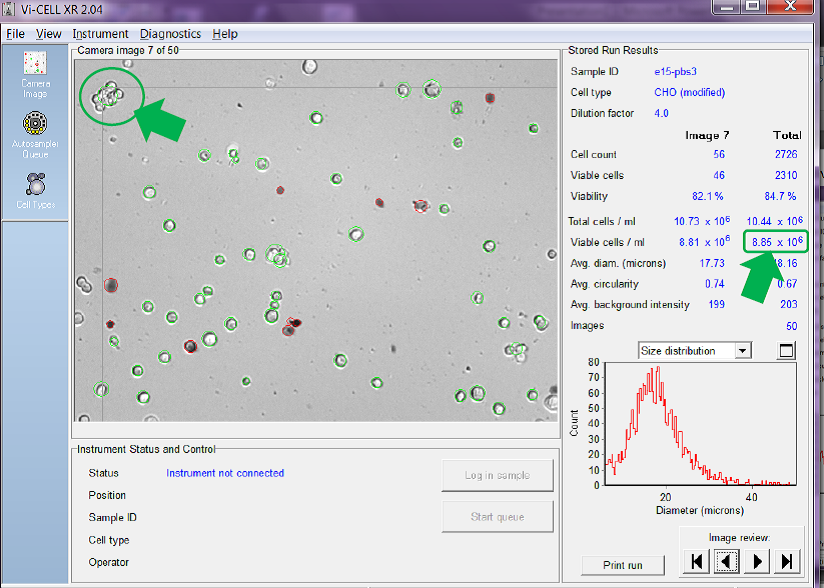
Figure 6. Counter set to count cells in clumps as one single cell
Obviously it depends on the number of cell clumps there are in the sample, but, in the example given above, counting cells in clumps instead of as individual cells reduces the reported total viable cell concentration from 10.53 x 106 cells/mL in Figure 5 to just 8.85 x 106 cells/mL in Figure 6, a reduction of circa 16%. This setting becomes even more important as the viable cell concentration increases and cells naturally become more ‘clumpy’.
A combination of the counter settings a) to g) allows the user to tailor the way the automated counter counts the cells so that it more closely mimics the way the user counts using the microscope, or is a closer match to the count results of an older counter that the new, automated counter is replacing.
Watch the webinar from Tony Harrison
Tony Harrison is a Product Manager and Compliance and Applications Specialist for Beckman Coulter Life Sciences. Tony has spent the last twelve years in applied metrology in the pharmaceutical and healthcare manufacturing industries. Prior to that, he worked for companies providing process control automation solutions for manufacturing industries.
Tony was joint-editor of the ISPE Guide to Ozone Sanitization of Pharmaceutical Water Systems4 and was also chief editor of the PHSS Best Practice Guide for Cleanroom Monitoring5.
Tony is a well-known international speaker and has provided educational seminars on TOC, liquid particle counting, ozone sanitization for water systems and cleanroom monitoring in UK, France, Italy, India, Germany, Malaysia, China, USA, Scandinavia, Ireland, Hungary, Switzerland, Indonesia, Belgium, Greece, Switzerland, Turkey, Egypt and Denmark.

