Leveraging Analytical Ultracentrifugation for Comprehensive Characterization of Lipid Nanoparticles in Drug Delivery Systems


Background
Analytical ultracentrifugation (AUC) is a well-established biophysical technique for the characterization of macromolecules. It has become the gold standard characterization method for adeno-associated viruses (AAVs) to determine particle loading states1 and has recently been applied to characterize lipid nanoparticles (LNPs) using sedimentation velocity and density contrast methods.2,3 Both methods provide information on the purity and comparability of different formulations. Density contrast AUC is used to experimentally determine the sample's density or partial specific volume distribution, enabling the characterization of unloaded particles, RNA copy number in the LNPs, and size distributions. AUC provides an array of unique information from a single technique, opens new possibilities, and enhances the characterization of LNPs for safer therapeutics.
Benefits of AUC:
- AUC is a first principle technique
- Due to the centrifugal force, AUC results in mixtures being fractionated during analysis, resulting in high-resolution results for heterogeneous samples
- AUC provides information about the size, shape and thermodynamic properties of particles
- With the ability to modulate speeds up to 60,000 rpm, AUC can characterize a wide range of particle sizes, from small peptides to large viral vectors
- The ability to select any wavelength between 190 to 800 nm allows for measurements of a wide range of concentrations and macromolecules
- A combination of hydrodynamic separation due to centrifugation and the collection of spectral properties allows for the characterization by orthogonal information
- AUC is an in-solution method that is non-destructive and requires no dyes or standards
Characteristics that need to be analyzed for LNP formulations:
- Percentage of unloaded LNPs in the sample
- Number of mRNA copies, or mRNA copy number distribution, that are loaded into the LNPs
- Size or hydrodynamic radius distribution
- Presence of aggregates or larger species
Introduction
Lipid nanoparticles (LNPs) are an advanced drug delivery system that encapsulates therapeutic molecules, such as nucleic acids, proteins, or small molecules, within a lipid-based particle.4 These particles typically consist of a core containing the active pharmaceutical ingredient (API), surrounded by a lipid shell. They can include cationic and ionizable lipids, which aid in encapsulating genetic material and facilitate its release inside target cells; phospholipids, which form the structural framework; cholesterol, which helps stabilize the lipid bilayer; and PEGylated lipids, which help increase stability and circulation time in the bloodstream4 (Figure 1). LNPs typically range in size from 20-100 nm, but the size can be modulated based on the amount of PEG used and the mixing.5
LNPs have gained significant attention for their role in delivering mRNA vaccines, such as those developed for COVID-19. LNPs offer several advantages, including increased stability, enhanced uptake by target cells, versatility for various applications, and general biocompatibility and biodegradability. They are being tested in multiple fields, including gene therapy, cancer therapy and protein delivery.6
Challenges still remain, however, in their manufacturing, scale-up, targeting specificity, and characterization. The challenges faced for the characterization of LNPs typically stem from their large size and inherent heterogeneity. As LNP research expands, several formulation characteristics are being investigated, including the percentage of unloaded particles, presence of aggregation or larger species, size or hydrodynamic radius distribution, and mRNA copy number distribution in the LNPs. Overall, lipid nanoparticles represent a promising and versatile platform for delivering a wide range of therapeutic agents, offering significant potential for advancing modern medicine.

 |
Ionizable neutral lipid |
 |
Ionizable cationic lipid |
 |
´Helper´ lipid |
 |
Cholesterol |
 |
PEG-lipid (polyethylene glycol lipid) |
 |
Nucleid acid (e.g. mRNA) |
Figure 1: LNPs are small particles used in the pharmaceutical and biotechnology industries to help improve drug delivery. They are composed of a lipids which encapsulate the nucleic acid or other therapeutic agent, allowing for improved cell targeting and enhanced drug efficiency.
The analytical ultracentrifuge (AUC) is used for the in-depth characterization of LNPs, as it provides unique insights into the size, mRNA loading state, and presence of unloaded particles in a single experiment. AUC is a first-principle, bulk analysis method that measures the samples as they are separated due to centrifugal forces based on their mass, shape and density. During centrifugation, several forces act on the particles: centrifugal forces cause macromolecules to sediment toward the bottom of the AUC cell, while frictional and buoyant forces—which oppose the centrifugal force-result in the diffusion of the macromolecules. Together, these forces create the sedimentation boundary that is monitored in AUC experiments (Figure 2). By analyzing sedimentation and diffusion behaviors of macromolecules in solution, it is possible to determine various physical properties of the molecules, such as their size, shape, mass and interactions.
Sedimentation velocity (SV)-AUC is one of the most common AUC methods; it has few buffer restrictions and can be used to determine purity and physical properties of particles and compare different samples or batches with each other.7 Density contrast AUC uses the principles of SV-AUC while measuring the sample multiple times in buffers that vary in density, which enables calculation of the density or partial specific volume of particles in the solution.3
Density contrast AUC is an attractive in-depth characterization approach for LNPs, as it results in the experimental determination of a density distribution, which can be used to calculate not only molar mass and size distribution but also RNA copy number distribution of the LNPs, and can help identify unloaded LNPs that might be present in the formulation.8 To help researchers implement these methods, we provide guidance below on the experimental setup and run recommendations for LNPs.
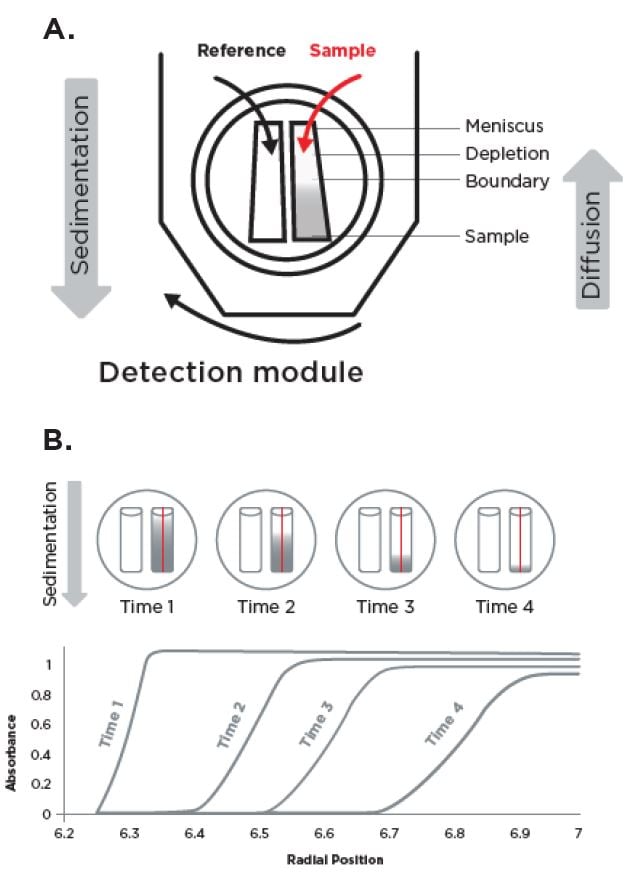
Figure 2: Schematic of an AUC experiment: A. Top view of an AUC cell assembly inside the rotor hole. B. Over the course of the experiment, detection modules provide a plot of sample concentration against radial distance along the sector length. Each time line is a scan that represents the sedimentation boundary of the sample throughout the experiment.
Protocol
1. Sample preparation:
a. Sedimentation velocity
Sample preparation for SV experiments is relatively easy, generally requiring only dilution or concentration of the sample to reach the desired concentration or optical density (OD) and selection of the measurement wavelength. The concentration range that can be accessed for experiments on the Optima AUC Analytical Ultracentrifuge is extensive, as any wavelength between 190 and 800 nm can be selected.
We recommend that users select a wavelength where the OD of the sample will be between 0.2 and 0.9 OD for the pathlength measured; for typical SV experiments with Beckman Coulter epon-charcoal centerpieces, this is 1.2 cm (cat no. Standard: 306493, flow-through: 392778). Once the sample is at the ideal concentration and the wavelength selected, the experiment can be set up and performed.2 If a sample is too highly concentrated, it’s possible that concentration-dependent non-ideality can occur, which results in non-ideal sedimentation patterns. Some software packages can help with modeling of these data; alternatively, if possible, measuring a diluted sample can help reduce or remove concentration-dependent non-ideality.9
b. Density Contrast AUC
For density contrast experiments, the same general sample preparation is followed. However, the sample needs to be measured at least four times in buffers that vary in density. For LNP studies, D2O or H2 18O can be used to create this variation in buffer density.2,3,8,10 We have created an Excel spreadsheet (LNP density contrast AUC sample prep) to aid users in creating these buffers, which can be found here. Users will require H2O, D2O, 20X buffer, and their concentrated stock sample.
Step 1: Determining the volume of stock LNP required: To determine the LNP volume required for AUC experiments, we recommend that you measure your stock sample using a spectrophotometer. The sample will most likely need to be diluted so that the absorbance at the wavelength you wish to perform the experiment at is below 1 OD. In the Excel spreadsheet, the dilution factor used should be added to Table 1 (Cell D5), and the corresponding OD for the wavelength of interest should also be recorded. We recommend repeating this measurement in triplicate, especially when using a nanodrop. Then, the dilution factor, the resulting OD, the total volume that will be used for the experiment (Cell D15), and the running OD (the OD you want to perform the experiment at; Cell E10) are used to determine the volume of sample required (Cell E11) for one experiment.
Step 2: Preparing samples in buffers that vary in their ratio of D2O and H2O: We recommend using at least four different D2O:H2O ratios; however, the exact ratios will depend on your formulation and its density. Samples should be prepared in buffers that maintain sample and salt concentrations but vary in their densities. Therefore, Table 2 in the LNP density contrast AUC sample prep Excel spreadsheet can aid in the calculation for preparations of the final solutions. If the D2O purity percentage varies from 99.9%, please update Cell D16. It should be noted that the density of LNPs loaded with RNA is typically close to water, and therefore, LNPs can sediment and float in the AUC. Figures 3A and 3B demonstrate what simulated data looks like for a sedimenting and floating particle, respectively.
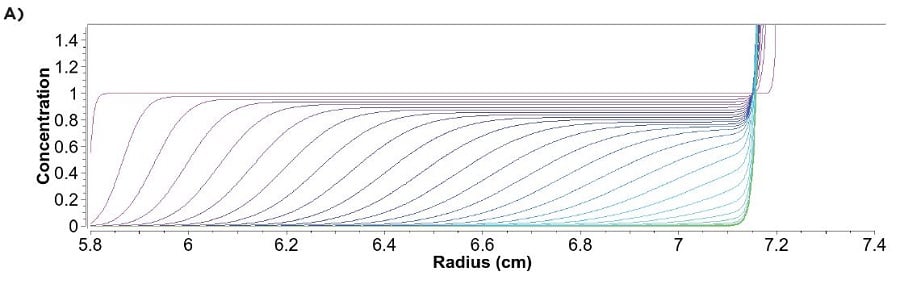

Figure 3: Simulation of sedimentation and floatation data: Simulated AUC data for a sedimenting (A) and floating (B) particle. The purple scans represent early scans; later scans are depicted in blue and green.
If an LNP formulation both sediments and floats in solution, a portion of the sample will have a sedimentation coefficient at or close to zero (Figure 4). Cases such as this should be avoided, as analytes with a sedimentation coefficient close to zero cannot be analyzed by the Lamm equation and, therefore, are not identified, and contribute only to the baseline and noise in the experiment.3 Consequently, it is recommended to use D2O:H2O ratios that result in the majority of the sample either sedimenting or floating. In the Excel spreadsheet, the percentages of D2O used can be updated by modifying cells D17-G17. Henrickson et al. used a wide range of D2O percentages in their study, ranging from 0% to 99%. This allowed for an extended range over which to extrapolate density, although they did report a couple of samples that crossed the zero sedimentation range, and required the use of two different speeds to measure samples.3 Bepperling and Richter used a range between 0 – 25% D2O, which allowed them to measure each of their 6 different samples in a single experiment.8
Figure 4: LNP-mRNA AUC experiment where the LNP density was close to the buffer density: A small amount of sedimentation and floatation can be observed for this sample; however, a majority of the sample has a sedimentation coefficient close to zero. This results in the boundaries rising upward over time rather than moving toward the right or left, as would be expected for a sedimenting or floating analyte, respectively.
2. Measuring the sample:
Once the sample preparation is complete, samples can be loaded into the AUC cells. Typical SV experiments are performed in 1.2 cm Beckman Coulter Epon Charcoal-Filled centerpieces (cat no. 306493) fitted with either sapphire (cat no. 392773) or quartz windows (cat no. 392772). When measuring wavelengths between 220-240 nm, use only sapphire windows purchased after 2020, or quartz windows (no purchase date exclusions).
There are two rotor options: the AN 50 Ti (cat no. 363782) for speeds ≤ 50 000 RPM, which has 8 cell positions for up to 16 samples, and the AN 60 Ti (cat no. 361964) rotor for speeds ≤ 60 000 RPM, which has 4 cell positions to measure up to 8 samples when measured in UV-Vis intensity mode.
The required speed to measure samples will vary depending on their size, mass, density and loading state. For RNA-loaded LNPs, speeds between 10,000 and 20,000 RPM have been used2,3,8,10. To determine if the selected speed is appropriate for your sample, the experiment should be monitored for the first 15 minutes after the rotor reaches the set speed and scan collection begins. If it is determined that the speed should be increased/decreased, the run can be stopped, and cells can be removed from the rotor and gently rotated for a couple of minutes to redistribute samples evenly. Samples and rotor can then be placed back in the instrument and the run can be restarted at the new speed.
We recommend collecting at least 25 scans/sample throughout the experiment, and that the sample be fully pelleted, either at the bottom or top of the cell, by the end of the experiment.
Analysis
After the run is completed, data can be exported from the AUC instrument and loaded into an analysis software. Recently, both UltraScan11 and SEDFIT12 have been used to analyze data demonstrating floatation and sedimentation.* Due to size and density of LNPs, they typically exhibit fast migration from sedimentation or floatation, which limits the resolution that can be obtained for their diffusion. This results in better resolution for the sedimentation coefficient and reduced resolution for the diffusion coefficient.13
It is important to note that the AUC data analysis assumes that the solvent's density and ionic strength are identical inside and outside the LNPs. While this assumption may hold true for dilute buffers like PBS, it could present issues in formulations with high concentrations of excipients, particularly salts, as they can alter the partial specific volume of the nucleic acid, potentially affecting the accuracy of the analysis.
Following is a short description of the two software packages, as well as information about what each software producer recommends for improving resolution of the diffusion coefficient to help improve the overall quality of an analysis.
UltraScan III11 supports use of custom grids to analyze samples that demonstrate sedimentation and floatation while excluding the 0 S region, where the Lamm equation fails.8,14 To overcome the reduced resolution obtained for the diffusion coefficient, it was recommended that the anisotropy (or f/f0) of your analysis be fixed to 1 or 1.1; the authors recommend that this be done if electron microscopy was used to identify the globularity of the LNPs.3 Using the custom grid method, the f/f0 can be fixed, and the sedimentation and partial specific volume values can be fitted. Additionally, UltraScan contains a Density Matching module that combines several analytical steps required to deduce distributions, such as partial specific volume, hydrodynamic radius, and molar mass from density contrast experiments. For more information on analysis of LNPs with UltraScan the user is directed to the publication by Henrickson, A., et al.3
SEDFIT also recently added a high-resolution, diffusion-deconvoluted sedimentation/floatation distribution analysis method.12 Their strategy to determine floatation coefficients and deconvolute the diffusion effects is to use a secondary method that can provide the hydrodynamic radius or diffusion coefficient, such as DLS, NTA, FFF, or EM. By providing the diffusion coefficient or hydrodynamic radius, the method can accurately deconvolute the floatation boundary and provide relatively high-resolution sedimentation coefficients for the floating material. They have demonstrated utility of the analysis for experimental models of extruded liposomes and for a commercial LNP product in their publication by Zhao, H., et al.12
mRNA copy number distribution determination
Density contrast AUC can be used to calculate the partial specific volume, which is the inverse of the particle’s density plus its hydration shell, and the molecular mass distributions of the LNP formulations; with these values, the mRNA copy number distribution can be calculated as described by Bepperling and Richter.8 This method can be used to determine the number of mRNA stands that are packaged into the LNPs; however, it should be noted that this method cannot differentiate between intact and degraded RNA. Briefly, this is done by determining the relative percentage of lipid in the LNP formulations, using equation 1:
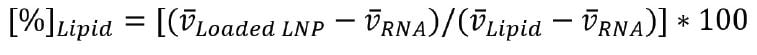
where v (Full LNP) represents each point on the partial specific volume distribution for the loaded LNP, v RNA is the partial specific volume of the RNA, and v Lipid is the partial specific volume of the empty LNPs. By subtracting the lipid percentage from 100, the relative percentage of RNA in the formulation can be obtained.
Values obtained for the percentage of RNA across the distribution can then be applied to the molecular mass distributions collected by AUC to determine the molecular mass contributed by the RNA at each point in the molecular mass distribution. Finally, by dividing these values through the theoretical molar mass of the RNA cargo, the distribution of the RNA copy number can be obtained.8
Conclusions
Analytical ultracentrifugation (AUC) is a powerful biophysical characterization technique that comprehensively characterizes LNP samples. Sedimentation velocity AUC has been used to characterize several different LNP formulations, demonstrating its strengths through the ability to detect multiple populations in polydisperse LNP-RNA formulations.10 Thaller et al. also studied the effect of different stressors on LNPs and found AUC to be more sensitive than DLS at detecting changes in the formulations.2 Density contrast AUC also results in the determination of mRNA copy number distributions, size distributions, and the detection of unloaded LNPs and encapsulation API.3,8,10,15,16 These studies highlight that AUC is a highly sensitive and accurate analytical technique for characterizing LNP formulations.
References
- Wang C, Mulagapati SHR, Chen Z, Du J, Zhao X, Xi G, Chen L, Linke T, Gao C, Schmelzer AE, Liu D. Developing an anion exchange chromatography assay for determining empty and full capsid contents in AAV6.2. Mol Ther Methods Clin Dev. 2019 Dec 13;15:257–263. PMCID: PMC6838793
- Thaller A, Schmauder L, Frieß W, Winter G, Menzen T, Hawe A, Richter K. SV-AUC as a stabilityindicating method for the characterization of mRNA-LNPs. Eur J Pharm Biopharm. 2023 Jan;182:152–156. PMCID: PMC9678208
- Henrickson A, Kulkarni JA, Zaifman J, Gorbet GE, Cullis PR, Demeler B. Density Matching Multiwavelength Analytical Ultracentrifugation to Measure Drug Loading of Lipid Nanoparticle Formulations. ACS Nano. 2021 Mar 23;15(3):5068–5076. PMID: 33617224
- Cullis PR, Hope MJ. Lipid nanoparticle systems for enabling gene therapies. Mol Ther. 2017 Jul 5;25(7):1467–1475. PMCID: PMC5498813
- Chen L, Ge B, Casale FP, Vasquez L, Kwan T, Garrido-Martín D, et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016 Nov 17;167(5):1398-1414.e24. PMCID: PMC5119954
- Mashima R, Takada S. Lipid nanoparticles: A novel gene delivery technique for clinical application. Curr Issues Mol Biol. 2022 Oct 19;44(10):5013–5027. PMCID: PMC9600891
- Zhao H, Brautigam CA, Ghirlando R, Schuck P. Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. Curr Protoc Protein Sci. 2013 Feb;Chapter 20:Unit20.12. PMCID: PMC3652391
- Bepperling A, Richter G. Determination of mRNA copy number in degradable lipid nanoparticles via density contrast analytical ultracentrifugation. Eur Biophys J. 2023 Jul;52(4–5):393–400. PMCID: PMC10248324
- Uttinger MJ, Walter J, Thajudeen T, Wawra SE, Peukert W. Brownian dynamics simulations of analytical ultracentrifugation experiments exhibiting hydrodynamic and thermodynamic nonideality. Nanoscale. 2017 Nov 23;9(45):17770–17780. PMID: 29131217
- Parot J, Mehn D, Jankevics H, Markova N, Carboni M, Olaisen C, Hoel AD, Sigfúsdóttir MS, Meier F, Drexel R, Vella G, McDonagh B, Hansen T, Bui H, Klinkenberg G, Visnes T, Gioria S, Urban-Lopez P, Prina-Mello A, Borgos SE, Caputo F, Calzolai L. Quality assessment of LNP-RNA therapeutics with orthogonal analytical techniques. J Control Release. 2024 Mar;367:385–401. PMID: 38253203



