Modular DNA Assembly of PIK3CA Using Acoustic Liquid Transfer in Nanoliter Volumes
Jared Bailey, Jefferson Lai, Rabia Khan, and John LesnickAbstract
Previous work has shown the ability of the Echo Liquid Handler to generate DNA constructs using various assembly chemistries with a miniaturized protocol1,2. Interchanging smaller, modular pieces of DNA is the preferred method for numerous workflows due to decreased synthesis cost and faster progression along the design, build, test, and learn cycle. In this study, NEBuilder HiFi was combined with the 25 nL increment dispensing Echo 525 Liquid Handler to assemble a modular oncogene construct encoding phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) over five modular pieces (Figure 2A). The modules themselves were designed to contain four activating mutations: R38H, E542K, E545K, and H1047R along with the wild-type PIK3CA in a pcDNA 3.1/Zeo(+) Vector for a plasmid size of 8,940 bp. The wild-type reaction was miniaturized down with a forty-fold reduction from the NEBuilder HiFi recommended volume in a previous experiment 2. At the fortyfold reduced volume, 1.25 fmoles of DNA was all that was required for each piece in a total reaction volume of 500 nL. The goal of this experiment was to utilize the Echo 525 Liquid Handler in assembling five, five-piece assemblies by substituting modular DNA pieces at the nanoliter scale. The experiment was validated by junction colony qPCR for rapid analysis of successful assembly and Illumina MiSeq sequencing verification for nucleotide resolution. The Echo Liquid Handler enables lower-cost methods and workflows to produce high-quality synthetic DNA constructs which expands design-based testing with higher throughput and affords the scientist a broader biological landscape to interrogate.
Introduction
Synthetic biology is an interdisciplinary science with the potential to impact academic and industrial applications including the creation of novel therapeutics and vaccines, plant science and biofuels, as well as bio-based chemical manufacturing capabilities that involve the application of engineering principles to biology. The focus is often on generating parts of natural biological systems, characterizing and isolating them, and then using them as components of an engineered biological system. A trademark of synthetic biology is the application of rational design principles to the design and assembly of biological components. The outcome of introducing a rationally designed synthetic DNA construct into a cell is difficult to predict. This creates the need to test multiple permutations to obtain the desired outcome. A greater emphasis on the modular design of DNA parts enables the assembly of a greater variety of potential constructs through combinatorial assembly of synthetic components. In addition to simplifying the overall DNA construction workflow, modular design and assembly of DNA components makes automation of the assembly process possible reducing the time, labor, and cost of generating multiple constructs to allow for an increase of throughput with an overall shortened development cycle.
Synthetic DNA plasmids are designed using computer-aided software with experimentally dependent functionality. This can range from interrogation of different domains of a single protein to the production of an entire pathway with heterologous genes. The designed DNA is then divided into synthesizable pieces. The pieces are designed with overlapping sequences to their neighbors before being chemically synthesized. The DNA fragments are then assembled together to build the designed construct using gene assembly techniques. If needed, multiple pieces can be assembled together into larger DNA assemblies. The assembled constructs are then typically cloned into an expression vector and sequence-verified. Once verified, the synthetic constructs are transferred into a production cell and the function of the designer construct is assayed. Depending on the results the constructs can then be modified or refined, and the test cycle is repeated until a DNA construct is obtained that produces the desired function.
New England Biolabs has developed the NEBuilder HiFi DNA Assembly Cloning Kit (Figure 1). The NEBuilder HiFi kit takes input DNA with 15 to 30 base pairs of terminal sequence identity and generates overhangs by using a proprietary 5’ exonuclease. The complementary 3’ overhangs are subsequently filled in with a DNA polymerase upon annealing. DNA ligase is used on the final product to seal nicks and create a continuous assembly. The entire reaction is incubated at a temperature of 50°C for as little as 15 minutes in a recommended volume of 20 μL.
Previous work has shown the ability of the Echo 550 Liquid Handler to generate two-piece assemblies using the Golden Gate and Gibson Assembly chemistries utilizing a miniaturized protocol 1. The Echo 550 Liquid Handler uses a transducer to acoustically dispense in increments of 2.5 nL. In another prior experiment, NEBuilder HiFi chemistry was combined with the 25 nL increment dispensing Echo 525 Liquid Handler to generate a two and five-piece assembly at the 500 nL volume2.
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) is a commonly studied gene in human oncology. The mutation of specific domains from this gene have been correlated with multiple tumor types 3–7. Due to PIK3CA’s importance in oncology, it was chosen to demonstrate a real-world application of an Echo 525 system-driven miniaturization of the NEBuilder HiFi kit. PIK3CA was divided into four modular pieces based on domains of interest 7 and was tagged with C-terminal GFP (Figure 2A). Four known oncogenic mutations, R38H, E542K, E545K, and H1047R were designed in modular pieces along with a wild-type control (Figure 2B).
The construct was also engineered to assemble into a KpnI-HF digested pcDNA 3.1/Zeo(+) Vector to allow for future transfection. The vector is interchangeably referred to as piece five after being linearized. The pieces were designed with 23 to 25 base pair DNA overlaps (Figure 2C), assembling five, five-piece assemblies by substituting modified modular DNA pieces at the nanoliter scale.
The Echo Liquid Handler enables lower-cost methods and workflows to produce high-quality synthetic DNA which expands design-based testing with higher throughput and affords the scientist a broader biological landscape to interrogate. Ultimately, the utilization of the Echo Liquid Handler for synthetic biology can help close the DNA read-write cost gap.
 |
 |
Figure 2.
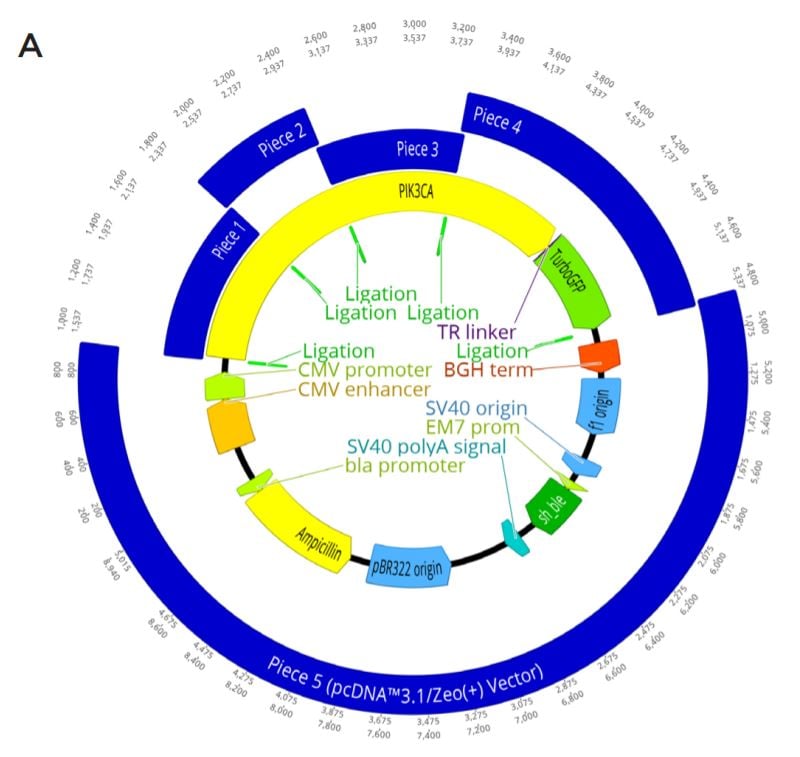
A. The complete plasmid construct to be assembled with GFP-tagged PIK3CA being generated in a KpnI-HF Digested pcDNA 3.1/ Zeo(+) backbone. gBlock Pieces ordered as well as linear vector are shown in blue, with their overlapping regions shown by a light green annotation labeled ligation. The PIK3CA gene is shown in yellow with the linker (purple) and turboGFP tag (green) also annotated. Other important parts of the plasmid machinery from pcDNA 3.1/Zeo(+) are also shown.
B. Nucleotide resolution for each of the designed overlaps in the assembly. This is enlarged from the Figure 2A ligation regions. Five overlaps were needed for the modular assembly.
C. The locations of the PIK3CA functional domains (dark green) and the planned mutations (red) within the constructs. The protein domains relationship with the synthetic piece design (blue) is also shown. The full length PIK3CA (yellow) gene is shown with its C-terminal GFP tag (light green). Each construct is designed to be either wild type or have one mutation. The planned mutations are R38H, E542K, E545K, and H1047R.
Modular DNA Assembly
Rational Contruct Design
A modular DNA assembly was also designed to be used in the miniaturized reaction volumes to more closely simulate the challenges faced in real-world applications. The PIK3CA cancer gene was chosen to be broken up into four pieces based on domains. These pieces were ordered as either wild-type sequence or containing the known oncogenic mutations, R38H, E542K, E545K, and H1047R. The assembly was also designed to insert into a linear pcDNA 3.1/Zeo(+) mammalian vector. This vector is interchangeably referred to as piece 5 after being linearized with KpnI-HF. All the pieces were designed with 23 to 25 base pair DNA overlaps and ordered from Integrated DNA Technologies as gBlocks Gene Fragments. The vector and gBlocks were amplified to obtain the necessary amount of modular DNA required for the study.
Modular DNA Component Production (gBlocks Pieces 1-4)
The primers listed in Table 1 were ordered from Integrated DNA Technologies (IDT) to generate additional copies of the modular DNA for assembly. The high-fidelity polymerase Q5 was used for the PCR reactions as shown in Table 2. Each reaction was set up in a 1.5 mL tube, gently mixed, and aliquoted into a 384-well Bio-Rad PCR plate. The reactions were then run on an Applied Biosystems ProFlex PCR System with the reaction conditions shown in Table 3.
gBLOCK Amplification Primers
| Piece # | Forward Primer Sequence | Reverse Primer Sequence |
| Piece 1-WT | TAGCGTTTAAACTTAAGCTTGGTACATGCCTC | AAGATGGCATTGTAAAACAGTCCATTGGC |
| Piece 1-R38H | TAGCGTTTAAACTTAAGCTTGGTACATGCCTC | AAGATGGCATTGTAAAACAGTCCATTGGC |
| Piece 2-WT | AGAAAGCCTTTATTCTCAACTGCCAATGGAC | CTGCGTGGGAATAGCTAAATCCTGCTTC |
| Piece 3-WT | GCAGGATTTAGCTATTCCCACGCAG | GACATGATGTCTGGGTTCTCCCAATTCAA |
| Piece 3-E542K | GCAGGATTTAGCTATTCCCACGCAG | GACATGATGTCTGGGTTCTCCCAATTCAA |
| Piece 3-E545K | GCAGGATTTAGCTATTCCCACGCAG | GACATGATGTCTGGGTTCTCCCAATTCAA |
| Piece 4-WT | GGACTAGTGGATCCGAGCTCGGTAC | ATTGGGAGAACCCAGACATCATGTCAGAG |
| Piece 4-H1047R | GGACTAGTGGATCCGAGCTCGGTAC | ATTGGGAGAACCCAGACATCATGTCAGAG |
Table 1. Primer sequences used to PCR amplify the gBlock template to increase concentration of necessary DNA for subsequent DNA assembly.
Piece PCR Reaction Conditions
|
Table 2. Volumes of reagents per PCR reaction used for template DNA expansion. The total reaction was dispensed in 20 μL aliquots into a 384-well Bio-Rad PCR plate. Q5 polymerase was chosen due to the need for a low error rate with the amplified pieces for downstream sequence fidelity. |
Thermocycler Reaction Conditions
| Piece # | Initial Denaturization | 25 Cycles | Final Extension | Hold | ||
| Time | 30 seconds | 10 seconds | 30 seconds | 1 minutes | 2 minutes | infinite |
| Temperature | 98oC | 98oC | 71oC | 72oC | 72oC | 4oC |
Table 3. Proflex thermocycler reaction conditions used for the amplification of the insert piece DNA (Pieces 1-4). The reactions were held to 25 cycles to minimize the potential to induce mutations.
Vector DNA Component Production (pcDNA 3.1/Zeo (+) Piece 5)
The circularized mammalian vector pcDNA 3.1/Zeo (+) was purchased from Thermo Fisher Scientific and transformed into NEB 10-beta competent E. coli. A clonal transformant was frozen in glycerol at -80°C for use in the entirety of the experiment. An overnight 25 mL culture was set up in a 250 mL Erlenmeyer Flask in LB Broth with 100 µg/mL carbenicillin and incubated at 37°C. It was split between 5 Qiaprep columns using the manufacturer recommended protocol and eluted in 250 μL. The eluted plasmid DNA was then digested overnight at 37°C with the addition of 20 μL NEB KpnI-HF and 30 μL 10x Fast Buffer.
DNA Cleanup, Quantitation, and Visualization (Pieces 1-5)
Upon reaction completion, both the vector restriction digest and gBlock PCRs were cleaned and concentrated using an NEB Monarch PCR & DNA Cleanup Kit (5 μg). The reactions were processed according to the manufacturer’s protocol and eluted in 11 μL of elution buffer.
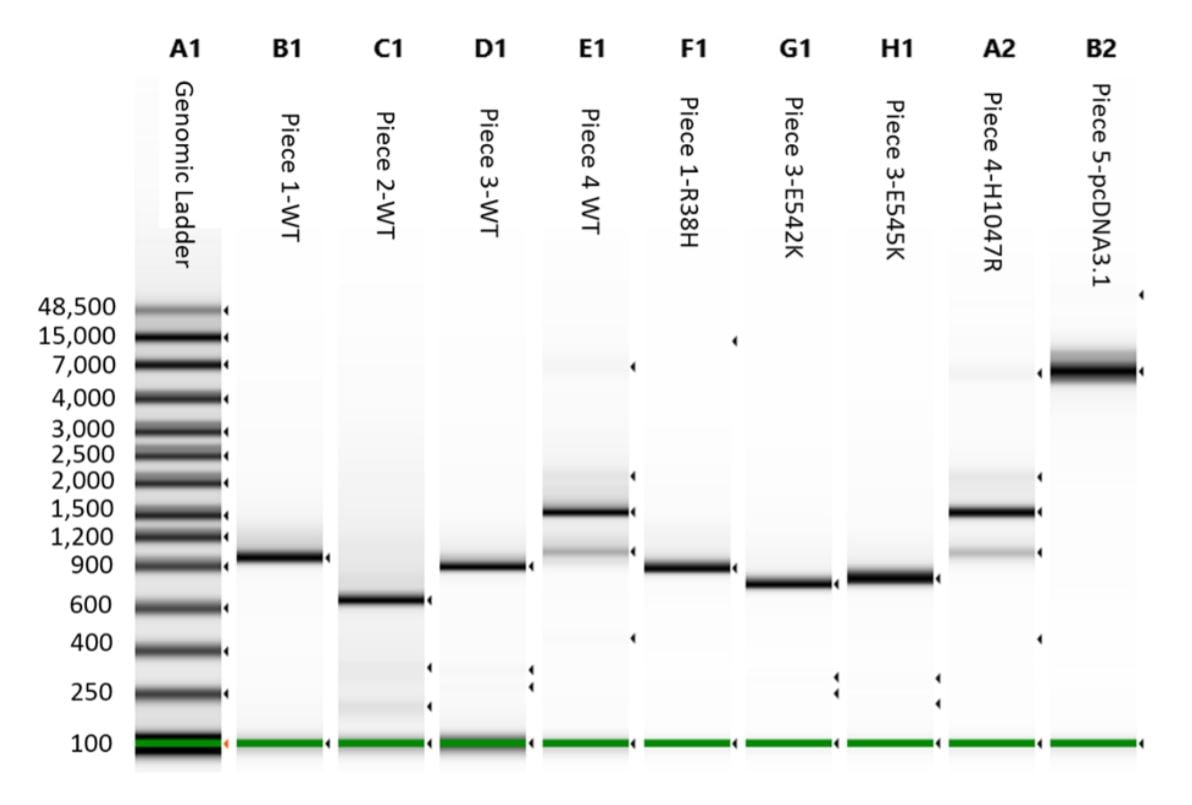
A 1 μL aliquot for each reaction was run on an Agilent 2200 Tapestation loaded with a Genomic DNA Analysis ScreenTape using their given protocol (Figure 3). The DNA was visually checked for quality, and the concentration of the expected band size was used as input for DNA normalization in DNA assembly.
Figure 3. Agilent 2200 Tapestation with Genomic tape run of the cleaned DNA assembly pieces. Tapestation concentration value for each correct band was used to calculate the proper reaction addition of each piece.
DNA and NEBuilder HiFi Master Mix Arraying
Using the DNA concentrations from the Agilent 2200 Tapestation run in Figure 3, the DNA was diluted to a desirable range for assembly (Table 4). The diluted DNA was then transferred into an Echo Qualified 384-Well Low Dead Volume Plus Microplate (384 LDV Plus). NEBuilder HiFi 2x Master Mix was also separately added to the 384 LDV Plus plate to allow for a low dead volume of enzyme master mix. The NEBuilder HiFi protocol has a recommended assembly volume of 20 μL with an input of 0.5 pmol per piece for complex assemblies. Prior work has demonstrated this reaction to be robust at the 500 nL volume with 1.25 fmol of each pieace. After arraying and sealing, the reaction was conducted for 1 hour at 50°C in an Applied Biosystems ProFlex PCR System with a heated lid. The PCR plate was cooled to 4°C before being transformed into NEB 10B E. coli cells.
NEBuilder Reaction Conditions
| Reagent | DNA Concentration (ng/µL) | ng DNA/1.25 fmol | WT Reaction (nL) | R38H Reaction (nL) | E542K Reaction (nL) | E545K Reaction (nL) | H1047R Reaction (nL) | Echo Calibration |
| Piece 1 WT | 23.6 | 16 | 25 | 25 | 25 | 25 |
384LDV_PIus_AQ_GP |
|
| Piece 1 R38H | 25 | 16 | 25 | |||||
| Piece 2 WT | 14.6 | 11 | 50 | 50 | 50 | 50 | 50 | |
| Piece 3 WT | 7.69 | 14 | 75 | 75 | 75 | |||
| Piece 3 E542K | 10 | 14 | 75 | |||||
| Piece 3 E545K | 10 | 14 | 75 | |||||
| Piece 4 WT | 29.6 | 26 | 50 | 50 | 50 | 50 | ||
| Piece 4 H1047R | 30 | 26 | 50 | |||||
| Piece 5 | 83.9 | 26 | 50 | 50 | 50 | 50 | 50 | |
| 250 | 250 | 250 | 250 | 250 |
- 1 hr 50oC Incubation (Reaction in Thermocycler with Heated Lid)
Table 4. Volumes of reagents used for the modular DNA assembly. The total reaction volumes were transferred by the Echo 525 Liquid Handler. These reagents were all dispensed from a 384 LDV Plus plate. These reactions were all transformed into E. coli.
Transformation/Construct Production
The modular DNA assemblies were transformed into 12.5 µL of chemically competent NEB 10B E. coli following the conditions outlined in Table 5. In addition to the five outlined DNA constructs, pUC19 was also transformed as a plasmid control. The volume of cells listed in the table was pipetted into the 384 PCR plate containing the reactions that had been chilled to 4°C. After a 30-minute incubation for the mixture at 4°C, the cells were heat shocked at 42°C for 30 seconds then immediately transferred back to the 4°C cold block.
Modular DNA E. Coli Transformations
| WT Reaction (nL) | R38H Reaction (nL) | E542K Reaction (nL) | E545K Reaction (nL) | H1047R Reaction (nL) | |
| NEB 10B cells | 12500 | 12500 | 12500 | 12500 | 12500 |
- 30 Minutes 4oC incubation (Cold Block)
- 30 Seconds 42oC (Heat Shock in Thermocycler)
- 2 minutes 4oC (Cold Block)
Table 5. Transformation conditions for NEB 10B E. coli cells into the various modular DNA assembly reactions. The cells were all added directly to the assembly reaction contained in the 384-well PCR plate
E. coli Plating
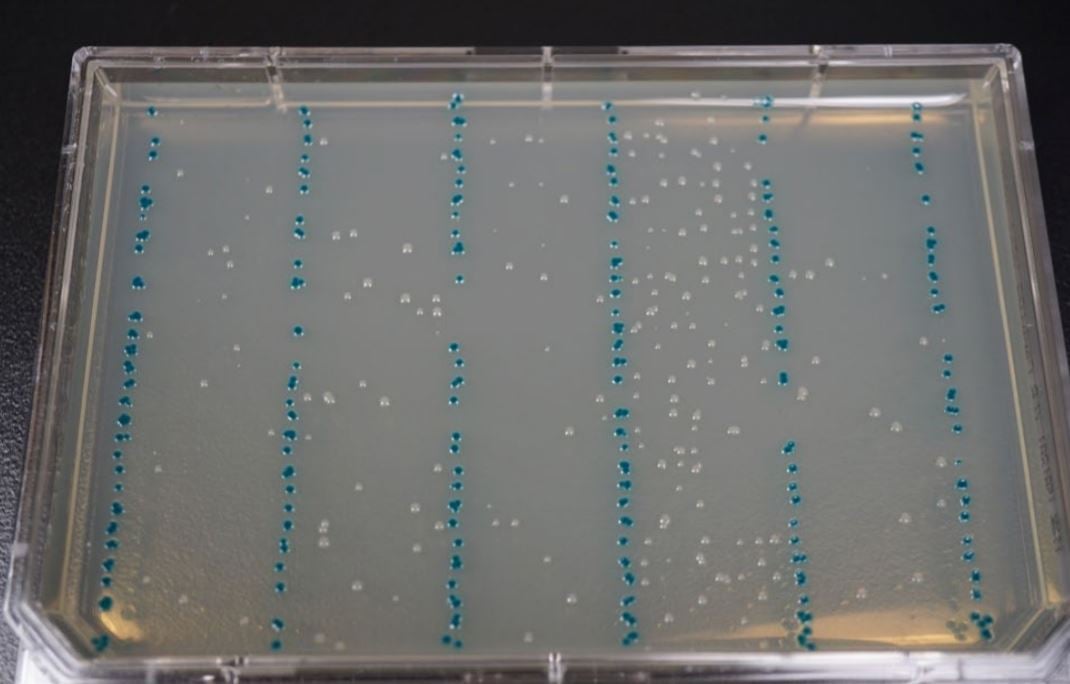
The transformed E. coli were manually transferred into a 384-Well LDV Plus plate after cooling and gently mixed by pipetting. LB Agar OmniTrays with 100 µg/mL carbenicillin, 60µg/ml X-Gal, and 0.1mM IPTG were warmed and dried in the 37°C incubator for 1 hour. The Echo calibration 384LDV_Plus_AQ_ GP was used to array 8 columns of 25 nL per construct transformation onto OmniTrays (Figure 4). Each experimental reaction was separated by a single 25 nL column of the pUC19 control. The plates were inverted and incubated overnight at 37°C. Isolated colonies were subsequently used for verification by colony qPCR and NGS. The resulting experimental colonies plates were used in both colony qPCR and NGS sequencing.
Figure 4. E. coli transformations spotted by an Echo 525 Liquid Handler onto an LB Agar OmniTray with 100 µg/mL carbenicillin, 60µg/ ml X-Gal, and 0.1mM IPTG. Each reaction was bracketed by a pUC19 control strain which is visible in blue. The pUC19 transformant contained an intact β-galactosidase pathway, which turns a blue color upon exposure to IPTG and X-Gal, so any cross-contamination between spots would be visible. From left to right, the reactions are: WT, R38H, E542K, E545K, and H1047R.
Colony qPCR Quality Control
The Echo 525 Liquid Handler was used to accurately dispense 250 nL of a cell suspension into four 10 μL qPCR reactions targeting four different junction regions of the assembled plasmid construct as a means of quality control for correct plasmid assembly (Figure 5). A qPCR reaction master mix was made for each of the four reactions containing the amplicon primer pair and LightCycler 480 SYBR Green I Master as shown in Table 6. The reagent mixture minus the colony suspension was bulk dispensed into a barcoded Bio-Rad Hard-Shell Skirted 384-Well PCR Plate. After reagent addition, the qPCR plate was centrifuged for 1 minute at 1000 × g. Isolated colonies were picked using a P10 pipette tip into 50 μL of diH20 in an Echo Qualified 384-Well Polypropylene Microplate 2.0 (384-Well PP). The cell-containing 384-Well PP plate was sealed and briefly mixed at 2000 RPM for 2 minutes on an Eppendorf MixMate. From this, 250 nL of the cell suspension was dispensed using the Echo calibration 384PP_AQ_BP into each of the four qPCR reagent mixes. The qPCR plate was then sealed with optical film, vortexed for 2 minutes at 2000 RPM on the Eppendorf MixMate, and briefly centrifuged at 1000 × g for 1 minute. After centrifugation, the sealed plate was run according to the conditions specified in Table 7. Colonies with all three junction CPs closely matched to the Ampicillin control were called positive assemblies and moved to Next-Generation Sequencing (Figure 6).
Colony qPCR Reaction Volumes
| Reagent | Volume (nL) per reaction | Echo Calibration |
| Forward Primer (10 µM) | 250 | |
| Reverse Primer (10 µM) | 250 | |
| LightCycler 480 SYBR Green I Master | 5000 | |
| diH20 | 4250 | |
| Colony Cell Suspension | 250 | 384PP_AQ_BP |
Table 6. Reagent additions used for the modular assembly colony qPCR reactions. Four different primer pairs were used to amplify the target regions from the colonies. Note that the Primers, SYBR Green Master Mix, and water were made as a master mix and dispensed together using a pipette. 250 nL of the cell suspension was then dispensed into each of the four different reaction conditions using the Echo 525 Liquid Handler.
Colony qPCR Reaction Conditions
| Temp. (oC) | Hold (sec) | ) Ramp Rate oC/sec | Acquisition (465 nm/520 nm) | Cycles | |
| Pre-incubation | 95 | 300 | 4.8 | 1 | |
| Amplification Piece 3 WT | 95 | 10 | 2.5 | 40 | |
| 50 | 10 | 2.5 | Single | ||
| 72 | 10 | 2.5 | |||
| Melting Curve Piece 3 E545K Piece 4 WT | 95 | 5 | 4.8 | 1 | |
| 65 | 60 | 2.5 | |||
| 97 | continuous | 0.11 | 5/oC | ||
| Cooling | 4 | 30 | 2.5 | 1 |
Table 7. Roche Lightcycler 480 conditions run for the DNA assembly junction qPCR looking for three generated junctions in comparison to the Ampicillin control.
Figure 6. Lightcycler 480 trace of a construct positive for all three experimental junctions in comparison to the Ampicillin control.
Miniaturized Nextera XT and MiSeq Plasmid Sequencing
The same cell suspensions used in the qPCR reaction were used by the Echo 525 system to spot onto an LB+Carb100 OmniTray. These spots were then expanded into 4, 96 deep well plates with 1.0 mL of LB+Carb100 liquid media. Each colony was set up in duplicate wells (2 mL media per colony) and incubated overnight at 37°C with shaking. The cell cultures were combined the next day and run through a Macherey-Nagel NucleoSpin 96 Plasmid Kit according to the manufacturer’s protocol. The DNA was eluted in diH20 and 50 μL transferred into an Echo Qualified 384-well PP plate. The QuantiT Picogreen dsDNA Assay Kit was used to quantify the amount of plasmid present in black walled Greiner 384-well polystyrene plates with a miniaturized protocol. First, 9 μL of 1xTE was added to all sample wells through a bulk dispense. The Echo 525 Liquid Handler was then used to add 1 μL of each DNA sample. The Echo 525 Liquid Handler also dispensed a standard curve in triplicate using the provided DNA standard. The curve ranged between 0.025 and 1 ng with a 10 μL final volume in TE. 10 μL of 1:200 diluted Picogreen dye was then added to all wells. The plate was sealed and mixed on the MixMate at 2000 rpm for 2 minutes before being centrifuged at 100 xg for 1 minute. The BMG Pherastar was used for the Picogreen quantitation with the excitation and emission peaks at 485 nm and 528 nm respectively. Based on these results, the DNA was normalized to provide 0.1 ng per tagmentation reaction. The Nextera XT tagmentation reaction and indexing reactions were then dispensed as shown in Table 8.
Nextera XT Reaction Conditions
| Tagmentation | Miniaturized (µL/rxn) | Echo Calibration |
| Plasmid DNA Sample | 0.5 | 384PP_AQ_BP |
| TD Buffer | 1 | 384PP_AQ_GP |
| ATM | 0.5 | 384PP_AQ_GPSB |
| Total volume | 2 |
- Spin @ 1500g, 1 minute -> Tagmentation reaction -> 55°C (5 min) --> 10°C (forever)
| Add NT | Miniaturized (µL/rxn) | Echo Calibration |
| Tagmentation reaction | 2 | From previous (no transfer) |
| NT Buffer | 0.5 | 384PP_AQ_SPHigh |
| Total volume | 2.5 |
- Spin @ 1500g, 1 minute -> Incubate 5 min RT
| Indexing Amplification | Miniaturized (µL/rxn) | Echo Calibration |
| Tagmentation reaction | 2.5 | From previous (no transfer) |
| Indexing Primer 1 (100µM) (N7XX) | 0.05 | 384PP_AQ_BP |
| Indexing Primer 2 (100µM) (55XX) | 0.05 | 384PP_AQ_BP |
| diH20 | 0.9 | 384PP_AQ_BP |
| NPM | 1.5 | 384PP_AQ_GPSB |
| Total volume | 5 |
PCR Reaction
| PCR Reaction | |
| 72°C | 3 min |
| 95°C | 30 sec |
| 95°C | 10 sec |
| 55°C | 30 sec |
| 72°C | 30 sec |
| 72°C | 5 min |
| 4°C | hold |
Table 8. Nextera XT Plasmid Tagmentation and Indexing
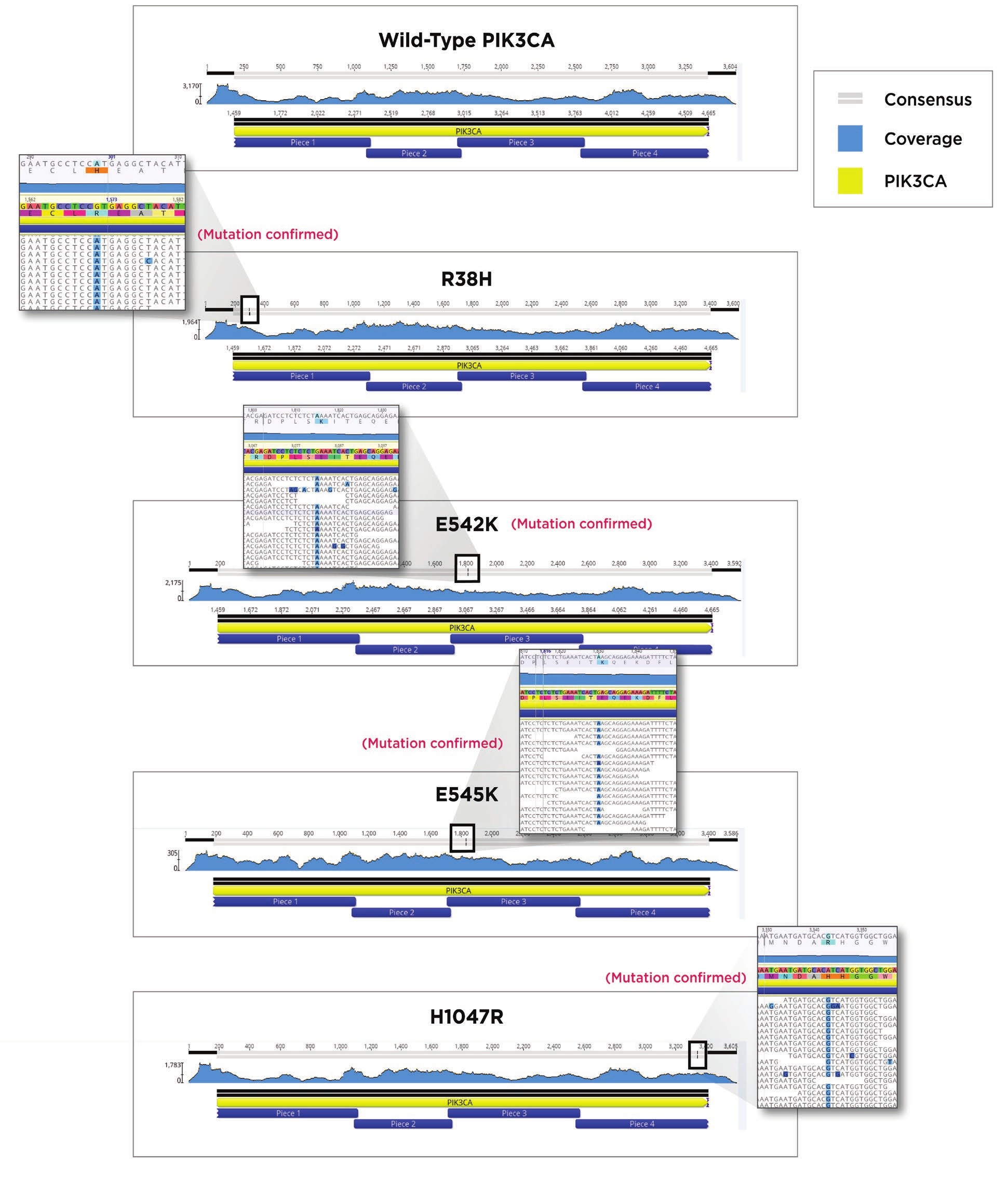
After indexing, Agencourt AmpureXP beads were used to clean up the reactions. The SPRI bead cleanup followed the Agencourt AmpureXP PCR purification protocol for 384-well format. We utilized 9μL of beads for our 5μL indexing amplification reaction, eluted in 20μL, and removed 15 μL with care not to carry over any beads. Quantitation was performed to verify the quantity of each sample using the Picogreen quantitation assay again as described above. Fragment size analysis was performed on the Agilent TapeStation 2200 with a TapeStation D1000 HS Kit, according to the Agilent standard protocol. Using the concentration data from the Picogreen assay, as well as fragment size information from the TapeStation 2200, we built a normalization worklist in Excel to obtain equimolar pooling of samples. The Echo 525 Liquid Handler was then used to simultaneously pool and normalize the libraries. The pool was then denatured and diluted to 20pM, loaded with 1% PhiX control, and run on an Illumina MiSeq specifying 2x75 reads. FASTQ files were generated on the MiSeq and aligned to an unmutated PIK3CA using Geneious 11.0. Correct PIK3CA constructs were obtained for each of the five desired assemblies as seen in Figure 7
Figure 7. MiSeq reads from each of the five constructs aligned to an unmutated PIK3CA gene. Sequence coverage and location of the mutation in the consensus read bar are shown at the top of each alignment. A magnified region containing the mutated amino acid residues are shown next to the four mutated versions of PIK3CA.
Discussion
Increasing the throughput of DNA construction is a goal of many research groups. The use of modular DNA pieces within a construction pipeline is a standard practice that offers the benefit of both lower synthesis cost as compared to a full-length gene and faster lead consolidation upon successful screening. The Echo 525 Liquid Handler, in combination with the NEB NEBuilder HiFi kit, can further enable this technique through the miniaturization of the reactions and workflow enhancements.
The use of a miniaturized 500 nL reaction volume while varying modular component pieces for a desired mutation was the primary aim of this experiment. The NEBuilder HiFi kit reaction volume was decreased forty-fold from the recommended volume from 20μL to 500 nL. At the 500 nL reduced volume, only 1.25 fmoles of DNA was used for each input piece, which would dramatically decrease the amount of DNA required from that of the recommended protocol which was 50 nmoles per piece. Obtaining sufficient high-quality input DNA is challenging, so the decrease of the required number of DNA molecules can vastly increase the number of modular assemblies possible. The use of these smaller volumes was done for PIK3CA constructs containing the mutations identified as R38H, E542K, E545K, and H1047R as well as wild-type in a mammalian expression vector. Correctly assembled clones were obtained for each of these targets as seen in the alignment from Figure 7. Successfully mutated PIK3CA constructs generated through the acoustic arraying of interchangeable modular pieces at nanoliter volumes alludes to a large scale increased-throughput DNA assembly workflow due to reduced reaction cost and DNA requirements.
Several other process improvements used in this study would help with an increase in the DNA assembly pipeline at larger scales. One such process improvement demonstrated in this experiment is the ability to acoustically plate the transformants onto solid media. OmniTray plating with the Echo 525 Liquid Handler can save operator time and incubator space as compared to traditional transformation plating. The Echo 525 Liquid Handler has technology that is uniquely positioned to make this plating possible. Tip-based systems have difficulty with poured agar plates being inherently concave and variable in height. The Echo 525 Liquid Handler is capable of transferring 25 nL increments of cell suspension very precisely onto the solid media on an X, Y grid, but with a buffered Z height due to the acoustic, tipless transfer of the droplet itself. This allows the transfer of cells onto solid media with minimal impact from the media height.
Another major process improvement was seen using colony qPCR for the transformed NEBuilder reactions. One of the major difficulties with any DNA construction technique is the removal of false positives, which can exceed 50% of clones upon Next-Generation Sequencing. Background vector is especially problematic whether it is re-circularized or uncut from the initial preparation. Using colony qPCR on the Echo 525 Liquid Handler, the background colonies can be removed prior to the timeconsuming plasmid recovery step. The colony qPCR is advantageous for scale-up when compared to end-point colony PCR due to the larger number of screened clones possible due to the removal of the laborious, low-throughput use of agarose gels. The quantitative results provided by the colony qPCR also remove the subjective nature of agarose gel reading. The colony qPCR was able to correctly predict 100% of vector background present, as well as 74% of the insertion deletion constructs from 192 samples sequenced allowing for a vastly enriched sequencing sample set. This process will be further optimized in future work. Finally, the sequencing of the passed plasmids can be done in a cost-effective way using a miniaturized Nextera XT protocol in conjunction with the Illumina MiSeq. Prior work had demonstrated the ability to miniaturize the Nextera XT kit ten to one hundredfold 8. In this experiment, tenfold Nextera XT reaction volumes were confirmed to also work for plasmid validation. Acoustic liquid transfers with the Echo 525 Liquid Handler allows the prospective user to decrease the overall reagent needs for NGS plasmid validation. Beyond overall DNA assembly reaction volume reduction, use of the Echo 525 Liquid Handler dramatically improves the DNA construction pipeline.
Working at the nanoliter scale in synthetic biology would have been inconceivable in the recent past, but the Echo Acoustic Technology makes these volumes a reality. Complex, modular DNA assemblies were generated using the NEBuilder HiFi kit at a forty-fold reduction in volume, which will significantly decrease the cost of DNA assembly allowing for substantial increases in throughput. The integration of a Echo 525 Liquid Handler into a DNA construction pipeline, such as one using the NEB NEBuilder HiFi kit, can allow for significant volume reductions as well as workflow enhancements with both complex and simple plasmid designs.
Materials
| Equipment | Manufacturer |
| Echo 525 Liquid Handler | Beckman Coulter Life Sciences |
| Allegra X-14 Centrifuge | Beckman Coulter Life Sciences |
| MixMate | Eppendorf |
| TapeStation 2200 | Agilent |
| Incu-Shaker Mini, Shaking Incubator | Benchmark Scientific |
| ProFlex PCR System | Thermo Fisher |
| LightCycler 480 | Roche |
| PHERAstar FS | BMG Labtech |
| Miseq | Illumina |
| Reagents | Manufacturer | Part Number |
| DNA Assembly Cloning Kit | New England BioLabs | #E5520 |
| KpnI-HF | New England BioLabs | #R3142 |
| Q5 High-Fidelity 2X Master Mix | New England BioLabs | #M0492 |
| QIAprep Spin Miniprep Kit | QIAGEN | #27104 |
| Genomic DNA Analysis ScreenTape Kit | Agilent | #5067-5365, #5067-5366 |
| NEB 10-beta Competent E. coli (High Efficiency) | New England BioLabs | #C2019l |
| T7 Express Competent E. coli (High Efficiency) | New England BioLabs | #C2566l |
| LB Broth with 100 µg/mL carbenicillin | Teknova | #L8185 |
| LB Agar OmniTrays with 100 µg/mL carbenicillin | Teknova | #L2010 |
| LB Agar OmniTrays with 100 µg/mL carbenicillin, 60µg/ml X-Gal, and 0.1mM IPTG | Teknova | #L2906 |
| gBlocks Gene Fragments | Integrated DNA Technologies C | Custom DNA |
| Junction qPCR primers | Integrated DNA Technologies C | Custom Oligos |
| Piece amplification primers | Integrated DNA Technologies C | Custom Oligos |
| LightCycler 480 SYBR Green I Master | Roche | #04707516001 |
| pcDNA 3.1/Zeo (+) Mammalian Expression Vector | Thermo Fisher Scientific | #V86020 |
| Monarch PCR & DNA Cleanup Kit (5 μg) | New England BioLabs | #T1030S |
| Nextera XT DNA 96-Sample Prep Kit | Illumina | #FC-131-1096 |
| Nextera XT Index Kit v2 Set A,B,C,D | Illumina | #FC-131-2001 |
| PhiX Control v3 | Illumina | #FC-110-3001 |
| MiSeq Reagent Kit v3 (150-cycle) | Illumina | #MS-102-3003 |
| Quant-iT Picogreen dsDNA Assay Kit | Thermo Fisher Scientific | #P11496 |
| Agencourt AMPure Beads | Beckman Coulter Life Sciences | #A63881 |
| 200 Proof Ethanol | Sigma Aldrich | #E7023 |
Materials (continued)
| Consumables | Manufacturer | Part Number |
| Echo Qualified 384-well PP Microplate | Beckman Coulter Life Sciences | #001-14555 |
| Echo Qualified 384-well LDV Plus Microplate | Beckman Coulter Life Sciences | #001-12782 |
| TapeStation Plate | Agilent | #5067-5150 |
| 14 mL Round Bottom Snap Cap Polypropylene Tubes | EK Scientific | #EK-62261 |
| Hard-Shell Skirted 384-Well PCR Plate | Bio-Rad | #HSP3805 |
| Hard-Shell 384-Well PCR Plates, Barcoded | Bio-Rad | #HSP3901 |
| Microseal 'C' PCR Plate Sealing Film | Bio-Rad | #MSC1001 |
| 384 well plates, polystyrene black (with micro-clear bottom) | Greiner | #781096 |
| Nalgene Single-Use PETG Erlenmeyer Flask (250 mL) | Thermo Fisher | #4112-0250 |
| 1.5 mL DNA LoBind Tubes | Eppendorf | #022431048 |