IOTest Myeloid Activation CD169-PE/HLA-DR-APC/CD64-PB Antibody Cocktail Analysis of Whole Blood [RUO]
Activation of Myeloid cells is an early response to infection. Myeloid cells of the innate immune system respond differently when challenged with bacteria or viruses1,2. A simple 3-marker combination gives an excellent overview of the entire innate immune response by analyzing peripheral blood.
Antibiotic resistance. Erroneous identification of source and nature of infection is a major challenge, leading to antibiotic misuse and resistance3. Rapid identification of bacterial infections, using nCD64, will aid research into biomarkers and development of new antibiotics.4
Viral disease progression. Recent research has shown that the interferon levels are modulated during the course of the disease. Type I IFN response, which increases in viral infections, such as COVID and HIV, becomes highly impaired in severe and critical subjects. Cellular markers such as monocyte CD169 are important research tools for understanding interferon activity during viral infection and disease progression.5,6,7,8
Interferon signaling. Type I and type II interferons (IFN) are central players in modulating the immune response. Recent evidence has demonstrated that these molecules regulate each other’s functions and that of other immune cells of the innate and adaptive immunity. Further research is underway to understand the role played by these molecules in infections, inflammatory diseases and cancer, especially as drug targets and biomarkers.8,9,10
Virus versus Bacterial Immune Response1, 2
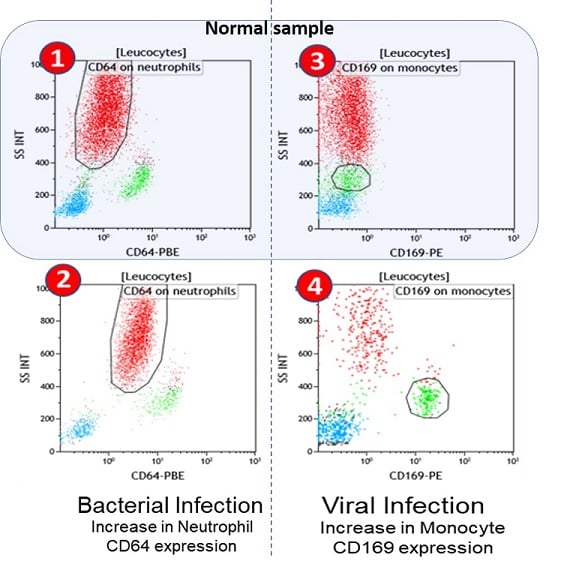
Flow cytometric plots representing the expression of CD64 (neutrophils) upon bacterial infection and CD169 (monocytes) upon viral infection.
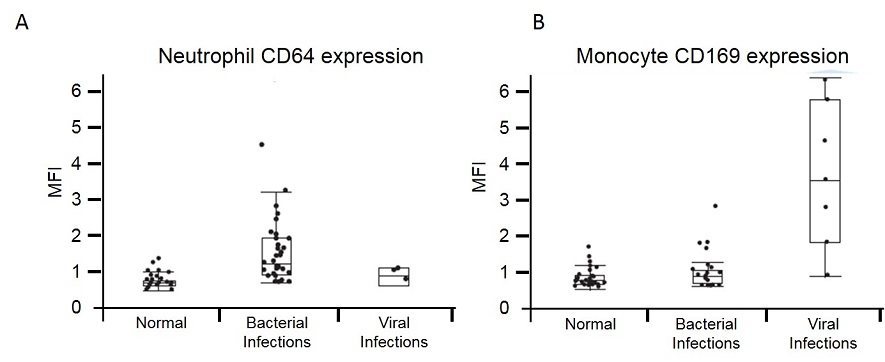
Neutrophil CD64 and Monocyte CD169 Levels for Three Groups. CD64 expression on neutrophils for Normal (no infection), Bacterial infections, and Viral infections, panel A. CD169 expression on monocytes for Normal (no infection), Bacterial infections, and Viral infections, panel B. Data are presented as mean and standard errors. The p-values are calculated between each group using student t test and reveal significant increase of biomarkers if p ≤ 0.05.
Staining in Whole Blood
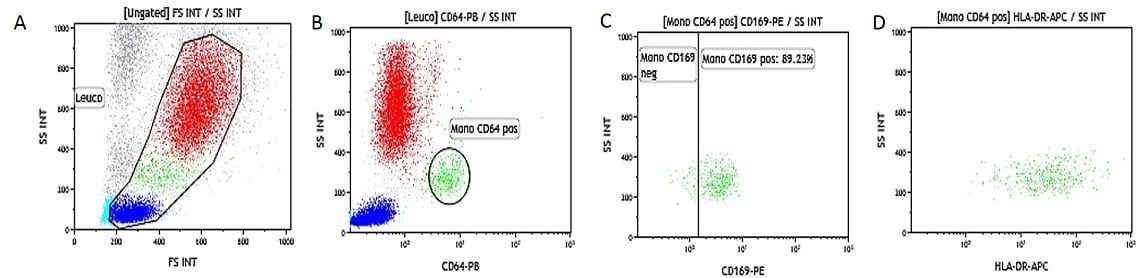
Heparinized Whole Blood IFNβ Activated. Data obtained with 10 μL of IFNβ activated whole blood (16-20 hour incubation at 37 °C with IFNβ at 0,1 μg/mL final concentration) and simultaneously stained and lysed according to the procedure described above. (A) SS/FS dot plot to gate Leukocytes, (B) SS/CD64-PB to gate monocytes gated on leukocytes, (C) SS/CD169-PE dot plot gated on monocytes (gate is placed on the non-activated control tube), (D) SS/HLA-DR-APC dot plot gated on monocytes.
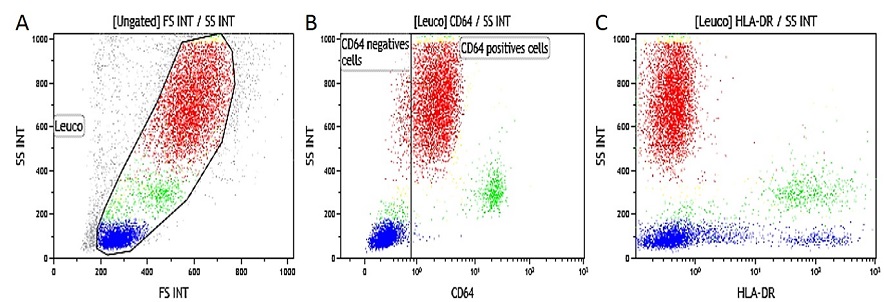
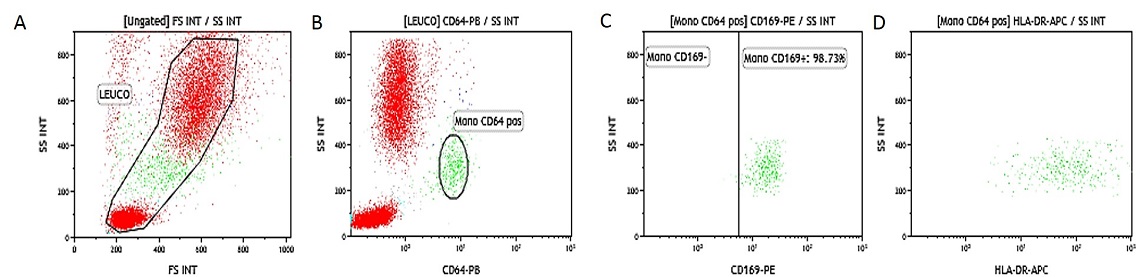
EDTA Whole Blood IFNγ Activated. Data obtained with 10 μL of IFNγ activated whole blood (16-20 hour incubation at 37 °C with IFNγ at 1 μg/mL final concentration) and simultaneously stained and lysed according to the procedure described above. (A) SS/FS dot plot to gate Leukocytes, (B) SS/CD64-PB gated on leukocytes (gate is placed on non-activated control tube), (C) SS/HLA-DR-APC dot plot gated on leukocytes.
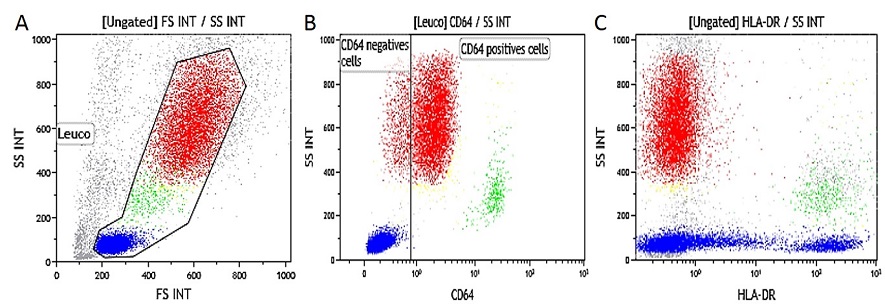
Heparinized Whole Blood IFNγ Activated. Data obtained with 10 μL of IFNγ activated whole blood (16-20 hour incubation at 37 °C with IFNγ at 1 μg/mL final concentration) and simultaneously stained and lysed according to the procedure described above. (A) SS/FS dot plot to gate Leukocytes, (B) SS/CD64-PB gated on leukocytes (gate is placed on non-activated control tube, (C) SS/HLA-DR-APC dot plot gated on leukocytes.
References:
- Bourgoin P, Biéchelé G, Ait Belkacem I, Morange PE, Malergue F. Role of the interferons in CD64 and CD169 expressions in whole blood: Relevance in the balance between viral- or bacterial-oriented immune responses. Immun Inflamm Dis. 2020;8(1):106-123.
- Bourgoin P, Soliveres T, Ahriz D, et al. Clinical research assessment by flow cytometry of biomarkers for infectious stratification in an Emergency Department. Biomark Med. 2019;13(16):1373-1386.
- Shallcross LJ, Davies DS. Antibiotic overuse: a key driver of antimicrobial resistance. Br J Gen Pract. 2014 Dec;64(629):604-5.
- Ajmani S, Agarwal V, Gurjar M. State of Globe: Neutrophil CD64: Is It a Reliable Biomarker for Sepsis? J Glob Infect Dis. 2018 Apr-Jun;10(2):33-34.
- Akiyama H, Ramirez NP, Gibson G, et al. Interferon-Inducible CD169/Siglec1 Attenuates Anti-HIV-1 Effects of Alpha Interferon. J Virol. 2017;91(21):e00972-17.
- Stegelmeier AA, van Vloten JP, Mould RC, Klafuric EM, Minott JA, Wootton SK, Bridle BW, Karimi K. Myeloid Cells during Viral Infections and Inflammation. Viruses. 2019 Feb 19;11(2):168.
- Bedin AS et al. Monocyte CD169 expression as a biomarker in the early diagnosis of COVID-19, medRxiv 2020.06.28.20141556.
- Park MD. Macrophages: a Trojan horse in COVID-19? Nat Rev Immunol. 2020 Jun;20(6):351.
- Hadjadj J, Yatim N, Barnabei L, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718-724.
- Rose T, Szelinski F, Lisney A, et al. SIGLEC1 is a biomarker of disease activity and indicates extraglandular manifestation in primary Sjögren's syndrome. RMD Open. 2016;2(2):e000292.
For Research Use Only. Not for use in diagnostic procedures.

