SPRIselect 磁珠试剂片段大小筛选方案
DNA size selection is a method used to isolate DNA fragments of a specific size range from a mixture of DNA fragments of varying lengths.
It is often employed in different molecular biology and genomic applications. For instance, DNA size selection is a crucial step of next-generation sequencing (NGS) library preparation because each NGS platform has an optimal DNA fragment size range that it can sequence most efficiently. Ensuring that the DNA fragments fall within this range helps maximize the sequencing data quality, accuracy and efficiency.
The protocol below describes procedures of left side, right side and double DNA size selections using our SPRIselect reagent powered by the SPRI bead-based technology. This protocol is interactive: you can specify the volume of your sample and the DNA fragment size range suitable for the NGS application and the platform you use, and the built-in calculator will instantly make all calculations for you.
NOTE: Some manufacturers suggest using our AMPure XP reagent for DNA size selection but we cannot guarantee performance or support this application. Only SPRIselect reagent was designed and validated for accurate and consistent DNA size selection from lot to lot. To better understand the difference between the AMPure XP and SPRIselect reagent, please visit this page.
- Magnet stand or plate
- 85% Ethanol, non-denatured
- Water (molecular biology grade) or a standard buffer solution such as Tris (10 mM, pH 8.0) or TE (10 mM Tris, pH 8.1 mM EDTA) for DNA elution
- Samples should be fragmented double-stranded DNA.
- Samples should be dissolved in molecular biology grade water or standard buffer solution such as Tris or TE.
- Sample volume is recommended to be ≥ 50 μL. A lower volume will decrease pipetting accuracy of the SPRIselect reagent, therefore increasing selection point variability.
- DNA fragments can be size selected in the range of 150–800 bp reliably.
- To maximize recovery for a left side size selection, the majority of the sample’s size distribution should be larger than the selection point.
- To maximize recovery for a right side size selection, the majority of the sample’s size distribution should be smaller than the selection point.
- To maximize recovery for a double size selection, the size distribution should be centered between the selection points.
This method eliminates DNA fragments shorter than the targeted size, enabling the selection of larger fragments.
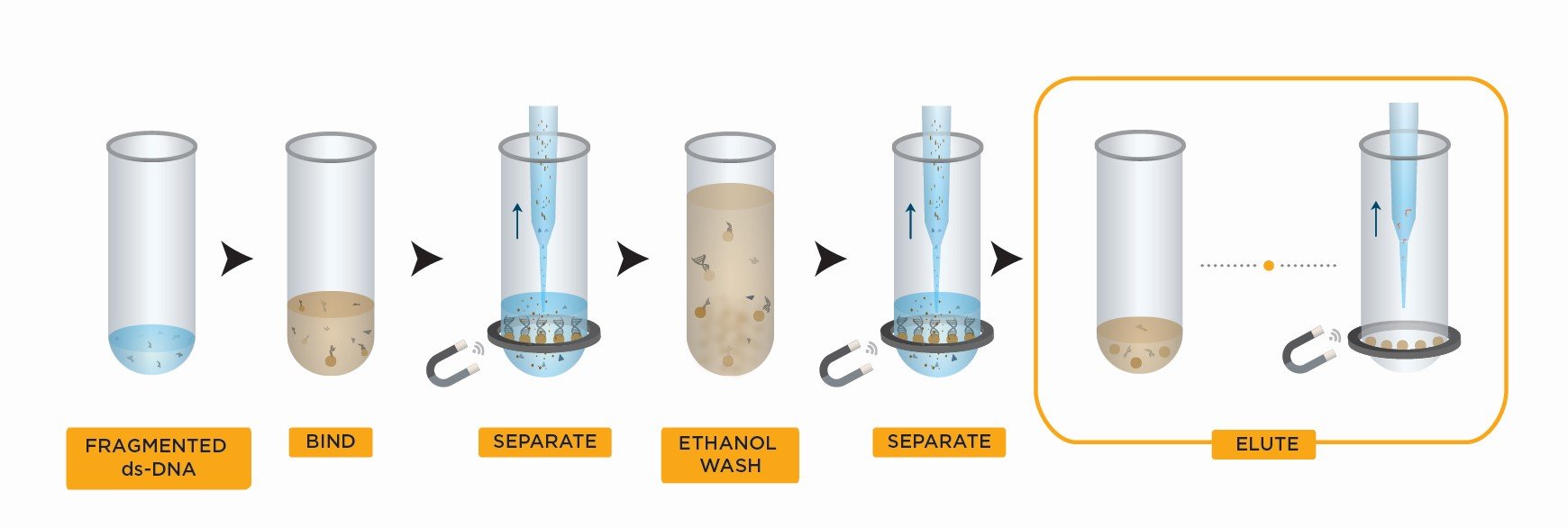
The Left Side Size Selection workflow starts with adding the SPRIselect reagent in your sample and mixing. As a result of this manipulation, the DNA fragments longer than the minimum targeted length get bound to the SPRI beads while the rest of DNA fragments stay floating in the solution.
The mix is then placed on a magnet for you to remove the supernatant (with the short DNA fragments we need to get rid of). Next steps are to wash the beads with ethanol and then elute in either water or a standard buffer solution.
As a general rule, increasing the ratio of the SPRIselect reagent volume to the sample volume (Bead Ratio) will increase the efficiency of selecting smaller fragments. The graphs below illustrate this relationship.
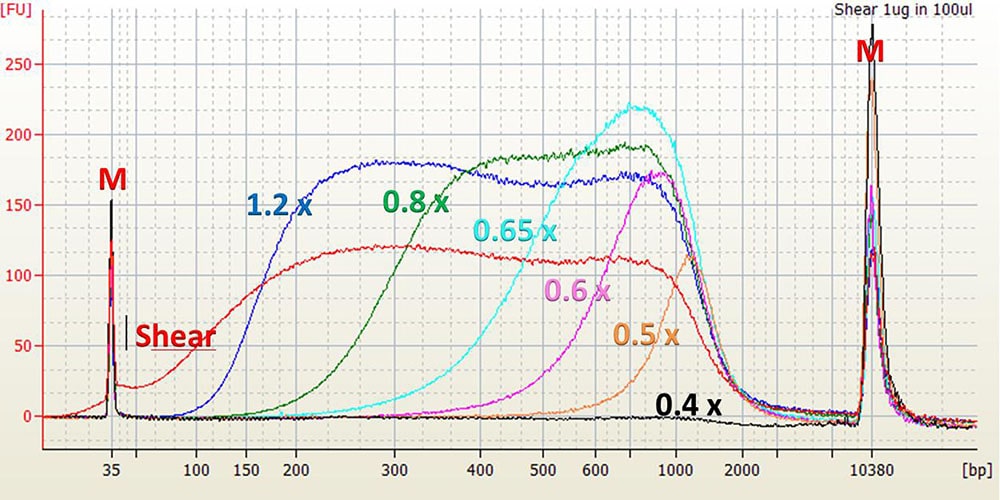
Agilent High Sensitivity DNA Chip Electropherogram. M = upper and lower markers for High Sensitivity DNA chip. Shear = 1 μL of 20 ng/μL input control sample in water. 1.2x to 0.4x = 1 μL of shear, size selected with given Bead Ratio.
Interactive Protocol
To adjust the Left Side Size Selection protocol to your conditions, please specify the volume of your sample and the minimum length of selected DNA fragments in the corresponding fields below. Our built‑in calculator will re-calculate the key protocol variables automatically.
KEY PROTOCOL VARIABLES CALCULATED
-
Thoroughly shake the SPRIselect bottle to resuspend the SPRI beads. Add μL of the SPRIselect reagent to μL of your sample.
CALCULATION
Formula: SPRIselect Volume = Sample Volume × Bead Ratio
For Your Conditions: SPRIselect Volume = μL × = μL
-
Mix the total reaction volume by pipetting 10 times and incubate at RT for 1 minute
OR
vortex for 1 minute at an appropriate speed until homogenous (depending on labware and total volume).NOTE: Insufficient mixing of the sample and SPRIselect reagent will lead to inconsistent size selection results. Make sure to mix well.
STEP RESULT: The DNA fragments of length bp are bound to the SPRIselect beads. The shorter DNA fragments float in the solution.
-
Place the reaction vessel on an appropriate magnetic stand or plate and allow the SPRI beads to settle to the magnet. Settle times will vary – a higher initial sample volume, a higher bead ratio or weaker magnets will require a longer settle time. Remove and discard the clear supernatant.
NOTE: Care should be taken not to aspirate more than a trace amount of beads during this step, as the desired library is associated with the beads. Significant bead loss will result in reduced yield.
STEP RESULT: The DNA fragments of length bp are bound to the SPRIselect beads. The shorter DNA fragments are removed.
-
With the reaction vessel still on the magnet, add 180 μL of 85% ethanol (non-denatured) and incubate at RT for 30 seconds. Remove and discard the ethanol supernatant.
NOTE: Care should be taken not to aspirate more than a trace amount of beads during this step, as the desired library is associated with the beads. Significant bead loss will result in reduced yield.
STEP RESULT: The SPRIselect beads holding the DNA fragments of length bp are washed.
-
To elute the sample:
-
Remove the reaction vessel from the magnet and add ≥ 20 μL of molecular biology grade water or standard buffer solution such as Tris or TE.
NOTE: Elution volume should be large enough so that the liquid level is high enough for the beads to settle to the magnet.
-
Mix the total elution volume by pipetting 10 times to resuspend the beads and incubate at RT for 1 minute
OR
vortex for 1 minute at an appropriate speed until homogenous (depending on labware and total volume). -
Place the reaction vessel on an appropriate magnetic stand or plate and allow the SPRI beads to settle to the magnet. Settle times will vary – a higher elution volume or weaker magnets will require a longer settle time.
-
-
Transfer the eluate (size selected sample) to an appropriate storage vessel.
STEP RESULT: The eluate containing the selected DNA fragments of length bp.
This method eliminates DNA fragments that exceed the targeted size, allowing for the selection of smaller fragments.
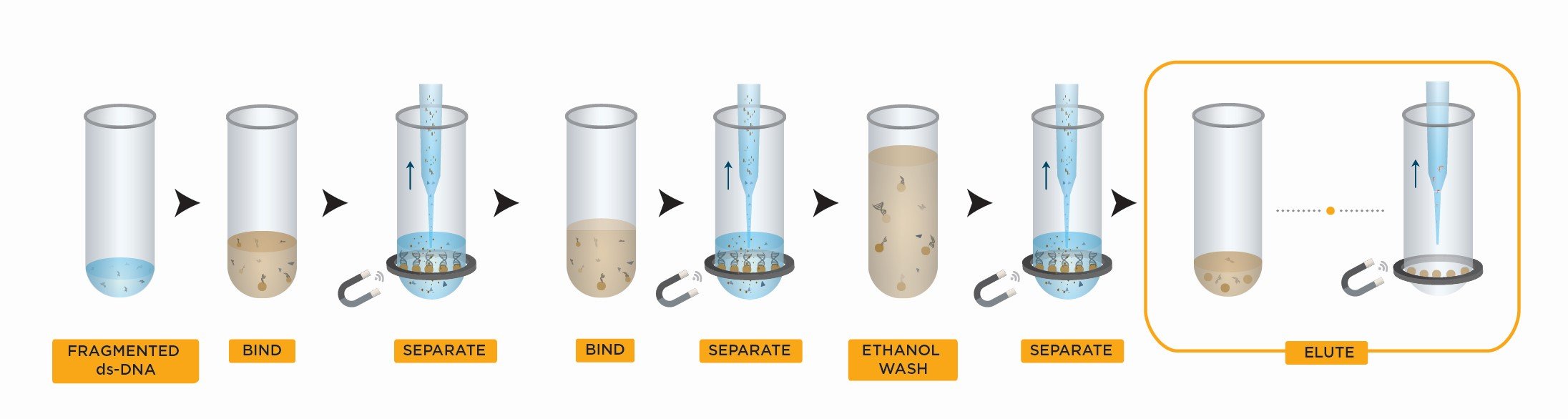
The Right Side Size Selection workflow is longer than the Left Side Size Selection workflow (see above). It starts with the same: adding the SPRIselect reagent in your sample, mixing and placing the mix on a magnet. But then, you should keep the supernatant (not the beads) to retain the DNA fragments shorter than the maximim targeted DNA size.
Next, you should add more SPRIselect reagent in this supernatant to perform left side size selection for getting rid of the unwanted shortest DNA fragments < 100 bp like primers, adapters and primer dimers. The last steps are to wash the beads with ethanol and then elute in either water or a standard buffer solution.
As a general rule, increasing the ratio of the SPRIselect reagent volume to the sample volume (Bead Ratio) will decrease the efficiency of selecting larger fragments. The graphs below illustrate this relationship.
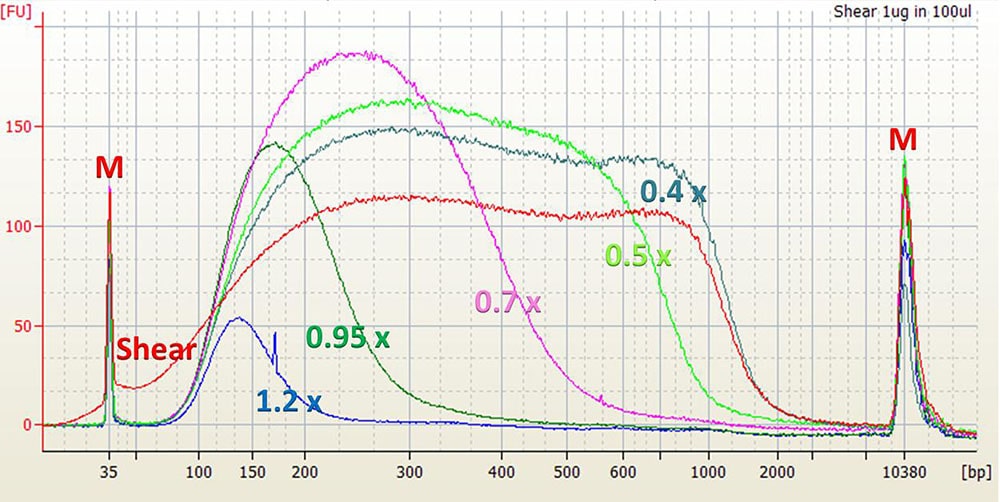
Agilent High Sensitivity DNA Chip Electropherogram. M = upper and lower markers for High Sensitivity DNA chip. Shear = 1 μL of 20 ng/μL input control sample in water. 1.2x to 0.4x = 1 μL of shear, size selected with given Bead Ratio.
Interactive Protocol
To adjust the Right Side Size Selection protocol to your conditions, please specify the volume of your sample and the minimum length of selected DNA fragments in the corresponding fields below. Our built‑in calculator will re-calculate the key protocol variables automatically.
KEY PROTOCOL VARIABLES CALCULATED
Thoroughly shake the SPRIselect bottle to resuspend the SPRI beads. Add μL of the SPRIselect reagent to μL of your sample.
CALCULATION
Formula: SPRIselect Volume 1 = Sample Volume × Bead Ratio
For Your Conditions: SPRIselect Volume 1 = μL × = μL
-
Mix the total reaction volume by pipetting 10 times and incubate at RT for 1 minute
OR
vortex for 1 minute at an appropriate speed until homogenous (depending on labware and total volume).NOTE: Insufficient mixing of the sample and SPRIselect reagent will lead to inconsistent size selection results. Make sure to mix well.
STEP RESULT: The DNA fragments of length bp float in the solution. The larger DNA fragments are bound to the SPRIselect beads.
-
Place the reaction vessel on an appropriate magnetic stand or plate and allow the SPRI beads to settle to the magnet. Settle times will vary – a higher initial sample volume, a higher bead ratio or weaker magnets will require a longer settle time.
-
Transfer the clear supernatant, which contains the DNA fragments of length bp, to a new reaction vessel. The reaction vessel with the remaining beads holding the DNA fragments of length > bp can be discarded.
NOTE: Care should be taken not to aspirate more than a trace amount of beads during this step, as the undesired larger fragment sizes are associated with the beads. Significant bead transfer will cause tailing into the larger size range.
-
Add μL of the SPRIselect reagent to the supernatant from Step 5 above.
CALCULATION
Formula: SPRIselect Volume 2 = Sample Volume × (1.8 - Bead Ratio)
For Your Conditions: SPRIselect Volume 2 = μL × (1.8 - ) = μL
STEP RESULT: The DNA fragments of length 100— bp are bound to the SPRIselect beads. The shortest fragments of length < 100 bp float in the solution.
-
Perform the following:
-
Mix the total reaction volume by pipetting 10 times and incubate at RT for 1 minute
OR
vortex for 1 minute at an appropriate speed until homogenous (depending on labware and total volume).NOTE: Insufficient mixing of the sample and the SPRIselect reagent will lead to inconsistent size selection results.
-
Place the reaction vessel on an appropriate magnetic stand or plate and allow the SPRI beads to settle to the magnet. Settle times will vary – a higher initial sample volume, a higher SPRIselect ratio or weaker magnets will require a longer settle time.
-
Remove and discard the clear supernatant.
NOTE: Care should be taken not to aspirate more than a trace amount of beads during this step, as the desired library is associated with the beads. Significant bead loss will result in reduced yield.
-
-
With the reaction vessel still on the magnet, add 180 μL of 85% ethanol (non-denatured) and incubate at RT for 30 seconds. Remove and discard the ethanol supernatant.
NOTE: Care should be taken not to aspirate more than a trace amount of beads during this step, as the desired library is associated with the beads. Significant bead loss will result in reduced yield.
STEP RESULT: The SPRIselect beads holding the DNA fragments of length 100— bp are washed.
-
To elute the sample:
-
Remove the reaction vessel from the magnet and add ≥ 20 μL of molecular biology grade water or standard buffer solution such as Tris or TE.
NOTE: Elution volume should be large enough so that the liquid level is high enough for the beads to settle to the magnet.
-
Mix the total elution volume by pipetting 10 times to resuspend the beads and incubate at RT for 1 minute
OR
vortex for 1 minute at an appropriate speed until homogenous (depending on labware and total volume). -
Place the reaction vessel on an appropriate magnetic stand or plate and allow the SPRI beads to settle to the magnet. Settle times will vary; a higher elution volume or weaker magnets will require a longer settle time.
-
-
Transfer the eluate (size selected sample) to an appropriate storage vessel.
STEP RESULT: The eluate containing the selected DNA fragments of length 100— bp.
Double Size Selection
This method selects DNA fragments within a specific size range.

The Double Size Selection workflow is the same as the Right Side Size Selection workflow (see above), just using different bead ratios. It starts with adding the SPRIselect reagent in your sample, mixing and placing the mix on a magnet. Then, you should keep the supernatant (not the beads) to retain the DNA fragments shorter than the maximim targeted DNA size.
Next, you should add more SPRIselect reagent in this supernatant to perform left side size selection for getting rid of the DNA fragments shorter than the minimum targeted DNA size. The last steps are to wash the beads with ethanol and then elute in either water or a standard buffer solution.
As a general rule, the ratio of the SPRIselect reagent volume to the sample volume (Bead Ratio) used for removal of long DNA fragments exceeding the targeted size range (Right Side Bead Ratio) always needs to be less than the bead ratio used for removal the short DNA fragments (Left Side Bead Ratio). The graphs below illustrate this relationship.
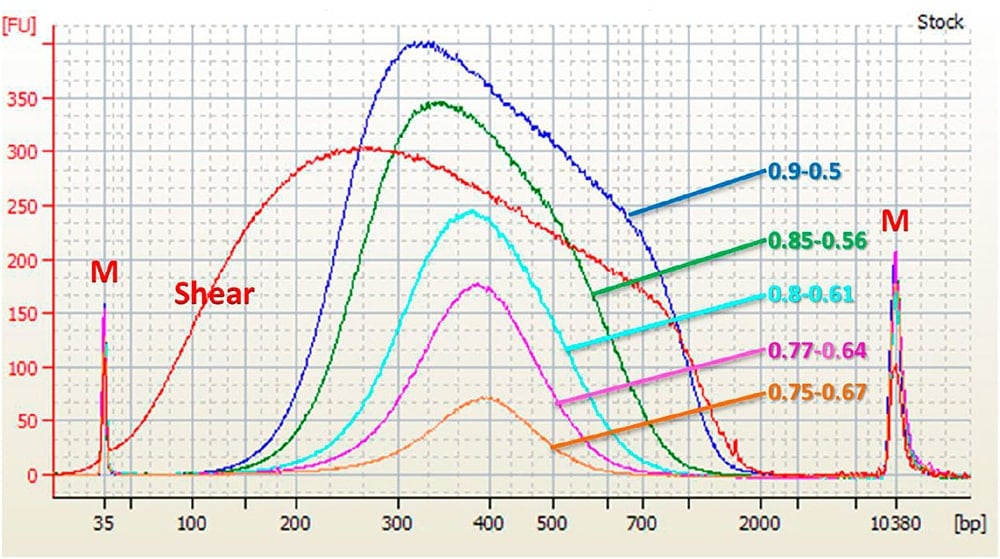
Agilent High Sensitivity DNA Chip Electropherogram. M = upper and lower markers for High Sensitivity DNA chip. Shear = 1 μL of 20 ng/μL input control sample in water. 0.XX to 0.XX = 1 μL of shear, size selected with given Left and Right Side Bead Ratio.
Interactive Protocol
To adjust the Right Side Size Selection protocol to your conditions, please specify the volume of your sample and the minimum length of selected DNA fragments in the corresponding fields below. Our built-in calculator will re-calculate the key protocol variables automatically.
KEY PROTOCOL VARIABLES CALCULATED
Thoroughly shake the SPRIselect bottle to resuspend the SPRI beads in it. Add μL of the SPRIselect reagent to μL of your sample.
CALCULATION
Formula: SPRIselect Volume 1 = Sample Volume × Right Side Bead Ratio
For Your Conditions: SPRIselect Volume 1 = μL × = μL
-
Mix the total reaction volume by pipetting 10 times and incubate at RT for 1 minute
OR
vortex for 1 minute at an appropriate speed until homogenous (depending on labware and total volume).NOTE: Insufficient mixing of the sample and SPRIselect reagent will lead to inconsistent size selection results. Make sure to mix well.
STEP RESULT: The DNA fragments of length < bp float in the solution. The larger DNA fragments are bound to the SPRIselect beads.
-
Place the reaction vessel on an appropriate magnetic stand or plate and allow the SPRI beads to settle to the magnet. Settle times will vary – a higher initial sample volume, a higher bead ratio or weaker magnets will require a longer settle time.
-
Transfer the clear supernatant, which contains the DNA fragments of length < bp, to a new reaction vessel. The reaction vessel with the remaining beads holding the DNA fragments of length > bp can be discarded.
NOTE: Care should be taken not to aspirate more than a trace amount of beads during this step, as the undesired larger fragment sizes are associated with the beads. Significant bead transfer will cause tailing into the larger size range.
-
Add μL of the SPRIselect reagent to the supernatant from Step 5 above.
CALCULATION
Formula: SPRIselect Volume 2 = Sample Volume × (Left Side Bead Ratio - Right Side Bead Ratio)
For Your Conditions: SPRIselect Volume 2 = μL × ( - ) = μL
STEP RESULT: The DNA fragments of length — bp are bound to the SPRIselect beads. The fragments of length < bp float in the solution.
-
Perform the following:
-
Mix the total reaction volume by pipetting 10 times and incubate at RT for 1 minute
OR
vortex for 1 minute at an appropriate speed until homogenous (depending on labware and total volume).NOTE: Insufficient mixing of the sample and the SPRIselect reagent will lead to inconsistent size selection results.
-
Place the reaction vessel on an appropriate magnetic stand or plate and allow the SPRI beads to settle to the magnet. Settle times will vary – a higher initial sample volume, a higher SPRIselect ratio or weaker magnets will require a longer settle time.
-
Remove and discard the clear supernatant.
NOTE: Care should be taken not to aspirate more than a trace amount of beads during this step, as the desired library is associated with the beads. Significant bead loss will result in reduced yield.
-
-
With the reaction vessel still on the magnet, add 180 μL of 85% ethanol (non-denatured) and incubate at RT for 30 seconds. Remove and discard the ethanol supernatant.
NOTE: Care should be taken not to aspirate more than a trace amount of beads during this step, as the desired library is associated with the beads. Significant bead loss will result in reduced yield.
STEP RESULT: The SPRIselect beads holding the DNA fragments of length bp are washed.
-
To elute the sample:
-
Remove the reaction vessel from the magnet and add ≥ 20 μL of molecular biology grade water or standard buffer solution such as Tris or TE.
NOTE: Elution volume should be large enough so that the liquid level is high enough for the beads to settle to the magnet.
-
Mix the total elution volume by pipetting 10 times to resuspend the beads and incubate at RT for 1 minute
OR
vortex for 1 minute at an appropriate speed until homogenous (depending on labware and total volume). -
Place the reaction vessel on an appropriate magnetic stand or plate and allow the SPRI beads to settle to the magnet. Settle times will vary; a higher elution volume or weaker magnets will require a longer settle time.
-
-
Transfer the eluate (size selected sample) to an appropriate storage vessel.
STEP RESULT: The eluate containing the selected DNA fragments of length bp.
Other Considerations
You should keep in mind that different additives in your sample like glycerol, MgCl2, polyethylene glycol, ethanol as well as factors like pH may influence the results of size selection (refer to the SPRIselect User Guide below for more details). Also, the size selection results depend on liquid volumes you handle. The smaller the volumes, the greater the variability. Therefore, we recommend adjusting the protocols above to your particular lab and workflow conditions while considering the volume values calculated in the protocols as starting points.
If you would like to decrease liquid handling variability or scale up the size selection workflow, you should think about automation. The described size selection protocols can be easily automated on liquid handlers like our Biomek i-Series Automated Workstation. Download the method overview below to learn more.
Still have questions? Contact our expert by completing the form.
Beckman Coulter makes no warranties of any kind whatsoever express or implied, with respect to this protocol, including but not limited to warranties of fitness for a particular purpose or merchantability or that the protocol is non-infringing. All warranties are expressly disclaimed. Your use of the method is solely at your own risk, without recourse to Beckman Coulter. Not intended or validated for use in the diagnosis of disease or other conditions. This protocol is for demonstration only, and is not validated by Beckman Coulter.


