分析型超速离心技术(AUC)在脂质纳米颗粒(LNP)表征中的应用综述
介绍
脂质纳米颗粒(LNP)和脂质体(图 1)作为多种治疗分子的递送载体,在医学领域展现出多维度突破性应用,已成功应用于癌症治疗、药物递送及疫苗开发,如 Moderna 和 Pfizer-BioNTech 推出的 COVID-19 mRNA 疫苗。由于 mRNA 具有免疫原性和毒性,且易被患者体内的核糖核酸酶(RNase)降解及肾脏过滤清除,因此不能以直接给药的方式注射至患者体内;1但通过将其包裹在脂质纳米颗粒(LNP)中,可成功克服上述问题。LNP 还可增强包裹药物的稳定性、实现靶向递送以及快速适配变异毒株等额外优势。2
LNP 的生物物理表征结果对其质量、功效和安全性评估至关重要。精准测定 LNP 制剂的粒径和均一性尤为关键,最新模型研究表明,这些参数直接影响治疗的免疫原性和效力。3 然而,LNP 制剂固有的异质性阻碍了粒径分布的精准测定。虽然动态光散射(DLS)是常用的粒径检测方法,但基于布朗运动的测量原理,其检测上限存在一定的局限性,可能导致聚集体漏检。4此外,DLS 检测法无法有效区分空壳和载药颗粒。为解决这一技术局限,美国 FDA 建议采用正交检测技术来验证表征结果。5 其他需表征的重要参数包括:游离载药和载体结合药物/包封药物、药物拷贝数分布、纳米颗粒的空载/满载比例(包封率)及其稳定性。

 |
可电离中性脂质 |
 |
可电离阳离子脂质 |
 |
“辅助”脂质 |
 |
胆固醇 |
 |
PEG 脂质(聚乙二醇脂质) |
 |
核酸(如 mRNA) |
图 1:脂质纳米颗粒(LNP)是医药和生物技术领域用于优化药物递送的纳米颗粒,其核心结构由脂质构成,能够有效包裹和保护核酸或其他治疗性药物分子,从而提升稳定性、细胞靶向性并增强药物效能。
分析型超速离心(AUC)技术是生物制药、疫苗研发等领域的重要工具,尤其在 LNP(脂质纳米颗粒)表征领域的应用日益广泛。在离心过程中,样品会基于其沉降系数(取决于分析物质量和密度)和扩散系数(取决于颗粒形状)发生流体力学分离。对于脂质纳米颗粒(LNP),其分离过程中的沉降或漂浮(图2),主要取决于脂质组分和载药量。通过实时追踪分析物在离心场中的光吸收特性,可测定其沉降/漂浮及扩散的动力学模式。基于测定的沉降和扩散系数,可进一步解析此类复杂体系的粒径分布、载药情况、摩尔质量等关键参数。
本文将探讨分析型超速离心(AUC)技术在脂质纳米颗粒(LNP)表征中的应用,并与其他分析方法进行对比。此外,现有研究表明,离心过程中产生的重力场似乎不会对 LNP 的结构或功能造成显著影响,反之,若存在此类影响,在分析过程中应能被精准识别。6
A)

B)

图 2:通过分析型超速离心机(AUC)收集的沉降和漂浮数据示例:AUC 离心过程中颗粒边界形状示例。Scan 图谱中的曲线颜色按扫描时间顺序编码,依次为紫色(最早)、蓝色(中期)和绿色(最新)。A)沉降颗粒的运动轨迹。B)离心过程中漂浮颗粒的运动轨迹。
一些研究使用分析型超速离心(AUC)技术来测定不同 LNP 制剂的粒径和粒径分布情况,包括 siRNA,mRNA 和阿霉素包封系统。5–7这些研究将 AUC 技术与其他技术在 LNP 平均粒径和粒径分布的检测结果进行比较研究,其他技术包括动态光散射(DLS)、纳米颗粒跟踪分析(NTA)、透射电子显微镜(TEM)和非对称流场流分离(AF4)与多角度光散射(MALS)。 研究发现,通过分析型超速离心(AUC)测定的平均粒径与其他所有测试方法的检测结果高度一致。值得注意的是,AUC 能精确测定所有 LNP 制剂的粒径分布,该结果与非对称流场流分离-多角度光散射联用技术(AF4-MALS)和冷冻透射电子显微镜(Cryo-TEM)技术的测定数据一致。AF4-MALS 在检测含有多分散性和高分子量物质的样品时,AF4-MALS 与 AUC 技术均展现出高分辨率检测优势5,8 (图 3),这得益于两种技术均结合了分离技术与实时在线监测能力。AUC 还额外增加了基于粒径和密度的双重分离维度,从而为 LNP 样品提供更精准的粒径分布测定结果。
此外,分析型超速离心(AUC)技术已被用于测定制剂中游离态与结合态载药的比例6,7 ,这一关键参数直接影响治疗安全性,因为游离载药可能引发毒性并增强免疫反应。9 Optima AUC 分析型超速离心机配备多波长光源系统,可在单次实验中实现 180-900 nm 范围内多达 20 个波长的同步检测。该技术能够实时监测制剂中不同载药的吸光度特性(如核酸在 240 nm 波长处,而阿霉素脂质体 Doxil 在 380 nm 波长处)。虽然 LNP 本身也可被检测,但由于脂质不吸收光,实际测得的是 LNP 产生的散射光信号,其有效检测范围通常在 215-280 nm 之间,具体取决于 LNP 的粒径。应注意:LNP 散射光信号的强度变化规律与载药的光吸收信号存在本质差异。6 通过在整个实验过程中测定的样品沉降和扩散系数,Mehn 等研究人员计算出了游离态药物的含量,其结果与 HPLC 和 DLS 技术的测定结果高度一致。7 Henrickson 等研究人员利用多波长与荧光检测技术证实其 siRNA LNP 仅含包封 RNA。6
A) 基于动态光散射(DLS)技术的批检测结果
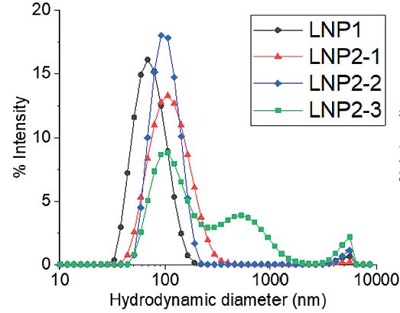
B) 基于多角度动态光散射(MADLS)技术的检测结果
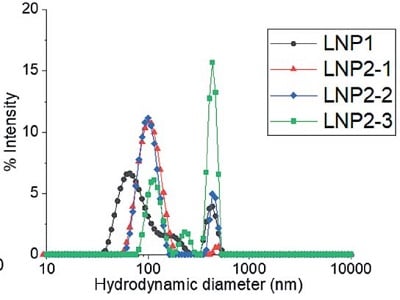
C) 基于纳米颗粒跟踪分析(NTA)技术的批检测结果
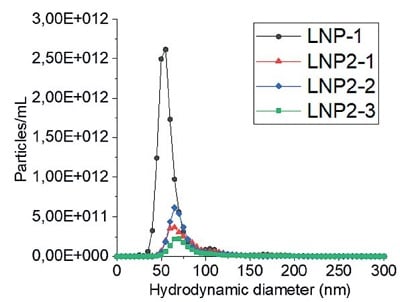
D) 基于分析型超速离心(AUC)技术的检测结果
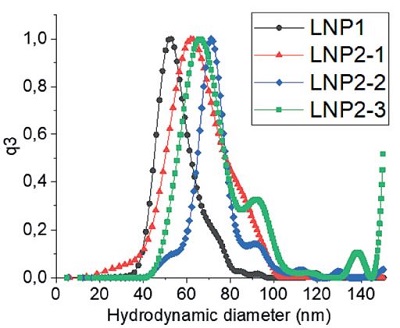
图 3:LNP 制剂的流体力学粒径:通过A)动态光散射(DLS)批检测、B)多角度动态光散射(MADLS)检测、C)纳米颗粒跟踪分析(NTA)批检测以及 D)分析型超速离心(AUC)技术测定的四种不同 LNP 制剂的流体力学粒径。有关图中参考颜色的更多信息和解释,请参阅 https://pubmed.ncbi. nlm.nih.gov/38253203/ Parot et al. DOI: 10.1016/j.jconrel.2024.01.037, Epub 2024, https://creativecommons.org/licenses/by/4.0/, image was not altered. 转载图片源文件,未作任何修改。
目前,尚不明确空载脂质纳米颗粒(LNP)在药物治疗过程中(若被使用)可能发挥的作用,但对其表征的研究有助于优化 LNP 生产工艺并确保治疗安全性。通过密度匹配分析超速离心技术(AUC),即在不同密度的缓冲液中多次测量同一样品,可测定样品的整体密度分布情况,获得该数据后,可将其与空载 LNP 样品的密度分布进行比较。若两者密度分布存在重叠区域,则表明该样品中可能含有一定比例的空载 LNP。5–7 Bepperling 和 Richter 基于该方法构建实验并计算出每个衣壳的 mRNA 拷贝数。10他们发现其 mRNA LNP 制剂的流体力学半径分布范围在 25 – 100 nm 之间,且每个衣壳包裹 1 – 10 个 mRNA 拷贝数。他们确定该个位数值的载药量与其粒径分布相匹配,且与同类 LNP 体系的研究数据一致。11–13 这些研究充分证实了分析型超速离心(AUC)技术在空壳 LNP 和组装完整的 LNP 分布及 mRNA 载荷定量表征方面的优势。这两项关键参数将直接影响细胞活性与 mRNA 表达动力学特征,12 对其精确测定不仅有助于优化 LNP 生产工艺,还能推动更广泛治疗药物的递送系统开发。需注意的是,在实际应用场景下(如冻融循环或室温操作等条件),必须通过多时间点监测来系统性评估 LNP 制剂的稳定性。9 Thaller 等研究人员对分析型超速离心(AUC)技术和动态光散射(DLS)在表征不同应力条件下 LNP 的多分散性和稳定性进行了比较。8DLS 技术可定性检测颗粒的流体力学半径,并识别暴露于冻融循环或室温条件下,颗粒在制剂中的变化以及在 50°C 条件下的机械应力(非热应力)。经研究验证,AUC 是一种卓越的 LNP 定量表征方法,可提供更精准的粒径分布结果,识别出颗粒在所有测试应力条件下的变化,并观察到使用 DLS 技术无法测定的颗粒密度变化。
这些研究强调了分析型超速离心(AUC)技术在表征 LNP 制剂领域的多功能性和实用性。AUC 可精准测定 LNP 制剂的粒径分布,且与非对称流场流分离-多角度光散射联用技术(AF4-MALS)和透射电子显微镜(TEM)技术的检测结果高度一致。此外,该方法还可成功识别和量化溶液中存在的游离载药和 空壳 LNP,并可用于确定每个 LNP 的 mRNA 拷贝数。总体而言,AUC 是一种无损的定量第一性原理方法,为 LNP 的表征提供了全面可靠的检测方法,已成为 LNP 研究中不可或缺的利器。
引用和参考文献
- Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014 Aug;15(8):541–55.
- Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, et al. SARSCoV- 2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020 Oct;586(7830):567–71.
- Chen L, Ge B, Casale FP, Vasquez L, Kwan T, Garrido-Martín D, et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016 Nov 17;167(5):1398-1414.e24.
- Caputo F, Vogel R, Savage J, Vella G, Law A, Della Camera G, et al. Measuring particle size distribution and mass concentration of nanoplastics and microplastics: addressing some analytical challenges in the sub-micron size range. J Colloid Interface Sci. 2021 Apr 15;588:401–17.
- Parot J, Mehn D, Jankevics H, Markova N, Carboni M, Olaisen C, et al. Quality assessment of LNP-RNA therapeutics with orthogonal analytical techniques. J Control Release. 2024 Mar;367:385–401.
- Henrickson A, Kulkarni JA, Zaifman J, Gorbet GE, Cullis PR, Demeler B. Density Matching Multi-wavelength Analytical Ultracentrifugation to Measure Drug Loading of Lipid Nanoparticle Formulations. ACS Nano. 2021 Mar 23;15(3):5068–76.
- Mehn D, Iavicoli P, Cabaleiro N, Borgos SE, Caputo F, Geiss O, et al. Analytical ultracentrifugation for analysis of doxorubicin loaded liposomes. Int J Pharm. 2017 May 15;523(1):320–6.
- Thaller A, Schmauder L, Frieß W, Winter G, Menzen T, Hawe A, et al. SV-AUC as a stability-indicating method for the characterization of mRNA-LNPs. Eur J Pharm Biopharm. 2023 Jan;182:152–6.
- Guerrini G, Magrì D, Gioria S, Medaglini D, Calzolai L. Characterization of nanoparticles-based vaccines for COVID-19. Nat Nanotechnol. 2022 Jun 16;17(6):570–6.
- Bepperling A, Richter G. Determination of mRNA copy number in degradable lipid nanoparticles via density contrast analytical ultracentrifugation. Eur Biophys J. 2023 Jul;52(4–5):393–400.
- Sabnis S, Kumarasinghe ES, Salerno T, Mihai C, Ketova T, Senn JJ, et al. A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates. Mol Ther. 2018 Jun 6;26(6):1509–19.
- Li S, Hu Y, Li A, Lin J, Hsieh K, Schneiderman Z, et al. Payload distribution and capacity of mRNA lipid nanoparticles. Nat Commun. 2022 Sep 23;13(1):5561.
- Carrasco MJ, Alishetty S, Alameh M-G, Said H, Wright L, Paige M, et al. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun Biol. 2021 Aug 11;4(1):956.


