Biomek i-Series Automated Illumina® Nextera XT DNA Library Prep Kit
Introduction
The Illumina Nextera XT DNA Library Prep Kit is used to prepare up to 96 indexed paired-end libraries for sequencing on Illumina platforms. The kit uses only 1ng of input DNA and fragments the DNA using an enzymatic reaction called Tagmentation. The kit saves time by providing master mixed reagents thus reducing pipetting optimizations and hands-on times. The kit accommodates various small genomes, amplicons, plasmids etc. The kit recommends bead based normalization, which can be effectively achieved in a fast and efficient way on Biomek automation and eliminates the need for quantification thus enabling higher throughput.
In this flyer, we demonstrate automated performance of Illumina Nextera XT DNA Library Prep Kit on the Biomek i7 Hybrid Genomics Workstation.
In comparison to manual pipetting, the Illumina Nextera XT DNA Library Prep Kit automated on Biomek platform provides:
- Reduced hands-on time and increased throughput
- Reduction in pipetting errors
- Standardized workflow for improved results
- Quick implementation with ready-toimplement methods
- Knowledgeable support
Spotlight: Biomek i7 Hybrid Genomics WorkstationSystem features deliver reliability and efficiency to increase user confidence and walk-away time.
|
Figure 1. Illumina Nextera XT DNA Library Prep Kit workflow
Automated Method
Automation provides increased efficiency, reducing hands-on time (Table 1). The method can be run start to finish with full walk away capability but does include optional start and stop points assigned based on Illumina’s recommendations, providing flexibility to the users in scheduling their workflow and breaking the process into pre and post steps.
| Major Process Descriptionn | Time | |
| 48 Samples | 96 Samples | |
| INSTRUMENT SETUP TIME PRE -PCR |
10 min | 15 min |
| Method Run Time POST -PCR |
51 min | 1 hr 29 min |
| Method Run Time | 1 hr 58 min | 2 hrs 14 min |
| Total | 3 hrs 34 min | 4 hrs 33 min |
*Timings estimates does not include thawing of reagent
**Method can be run with full walk-away capability after initial setup
Table 1. Estimated run times for Illumina Nextera XT DNA Library Prep Kit on the Biomek i7 Hybrid Genomics Workstation.
The software provides several user friendly features such as:
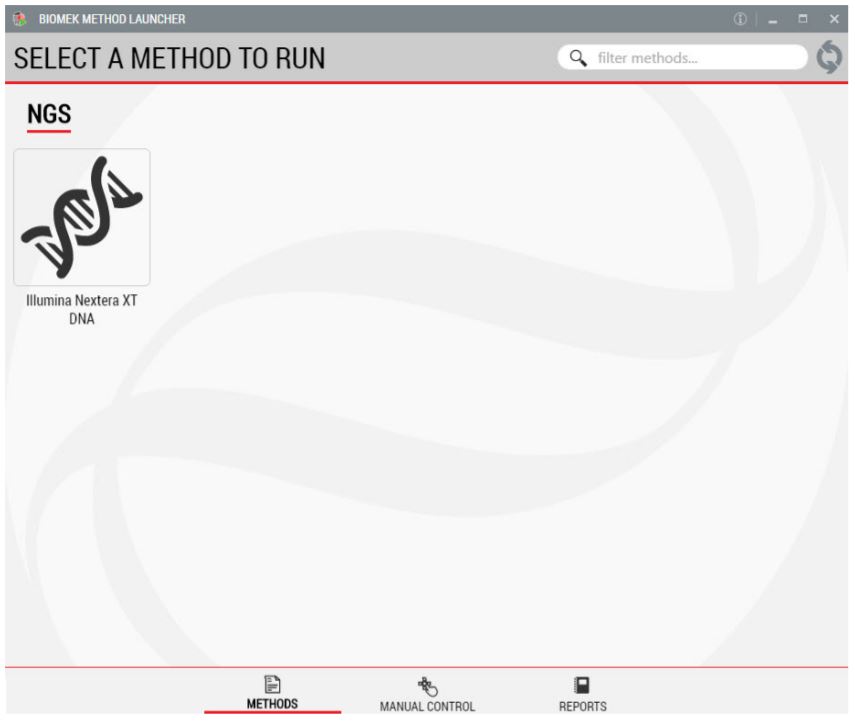
1. Biomek Method Launcher (BML)
BML is a secure interface for method implementation without affecting method integrity. It allows the users to remotely monitor the progress of the run. The manual control options provide the opportunity to interact with the instrument, if needed.
Figure 2. Biomek Method Launcher provided an easy interface to start the method.
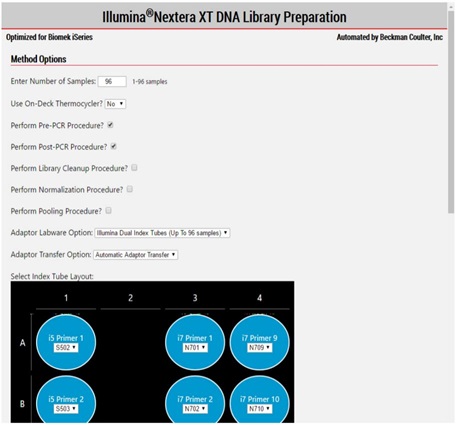
2. Method Options Selector (MOS)
MOS enables selection of plate processing and sample number options to maximize flexibility, adaptability and the ease of method execution.
Figure 3. Biomek Method Options Selector enables us to select sample number and on or off deck Thermocycler option and all the processing options.
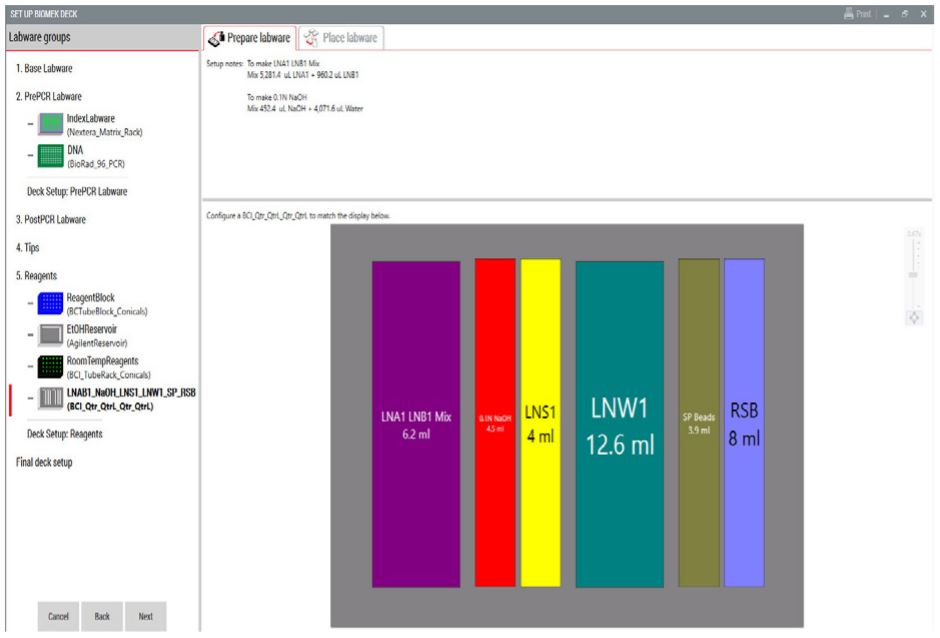
Figure 4. Guided Labware Setup indicates reagent volumes and guides the user for correct deck setup.
3. Guided Labware Setup (GLS)
GLS is generated based on options selected in the MOS, and provides the user specific graphical setup instructions with reagent volume calculation and step by step instructions to prepare reagents.
Experimental Design
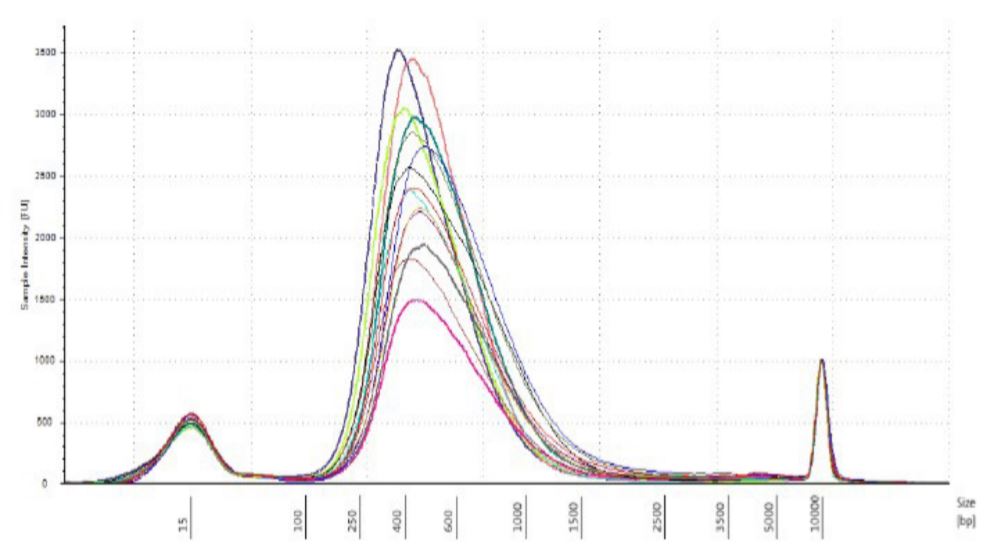
Eight technical replicates of genomic DNA of each of the ATCC strains (Bacillus Cereus ATCC strains 14579 D-5, Escherichia Coli 700926 D-5, and Rhodobacter Sphaeroides 17023 D-5 respectively) were used for this experiment. 1ng (0.2ng/uL) of input DNA was used per well to generate libraries with Nextera XT DNA kit. The quality of the libraries was assessed using Agilent Tapestation 2200 and High Sensitivity D5000 tape. The libraries were then sequenced on the MiSeq using the MiSeq Reagent Kit v3 600-cycle.
Results
A robust and reliable automation solution for NGS library prep is essential to achieve reduced error and to take full advantage of Illumina’s sequencing technology. Automation saves up valuable time for researchers.
|
Figure 5. DNA strains analyzed on TapeStation 2200 with High Sensitivity DNA 5000 tape. |
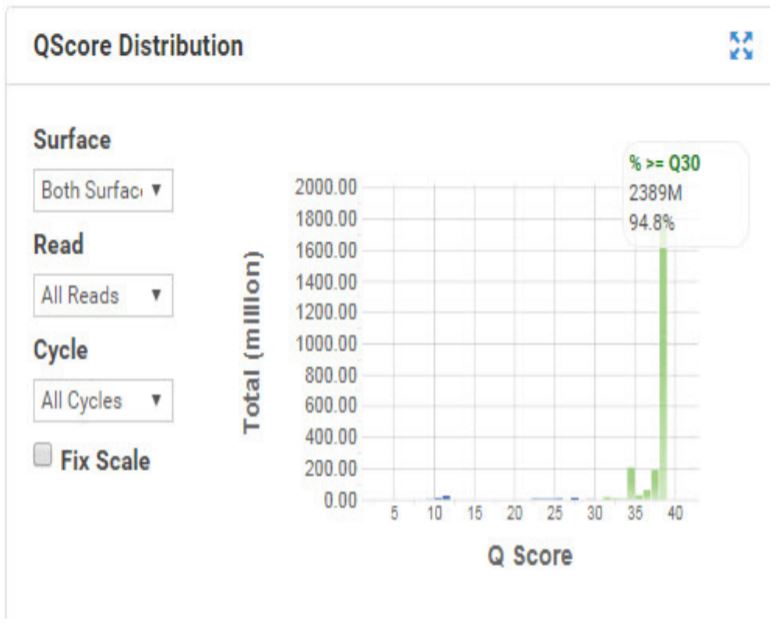
Figure 6. Q Score distribution for the HiSeq Run for Nextera XT DNA. |
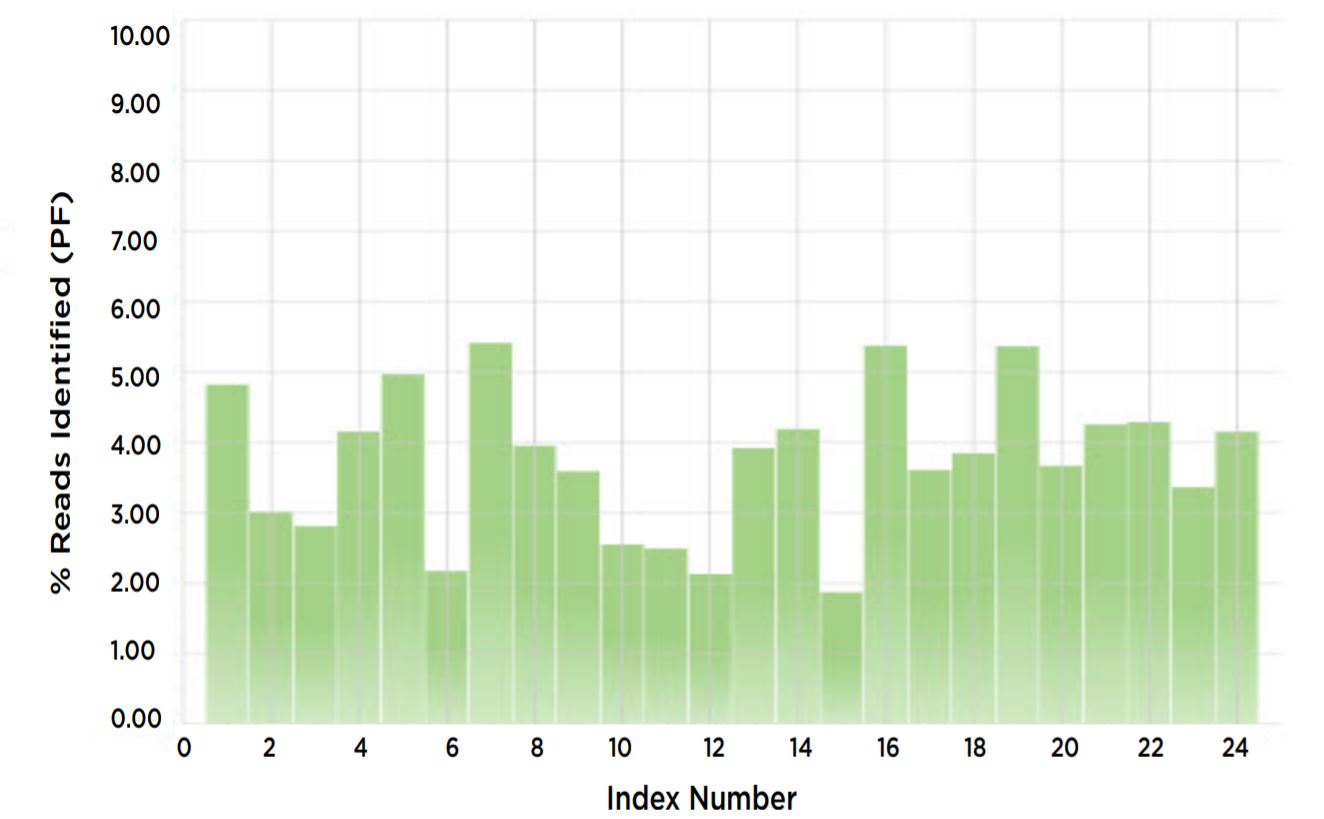
Figure 7. % reads identified for each index proportional.
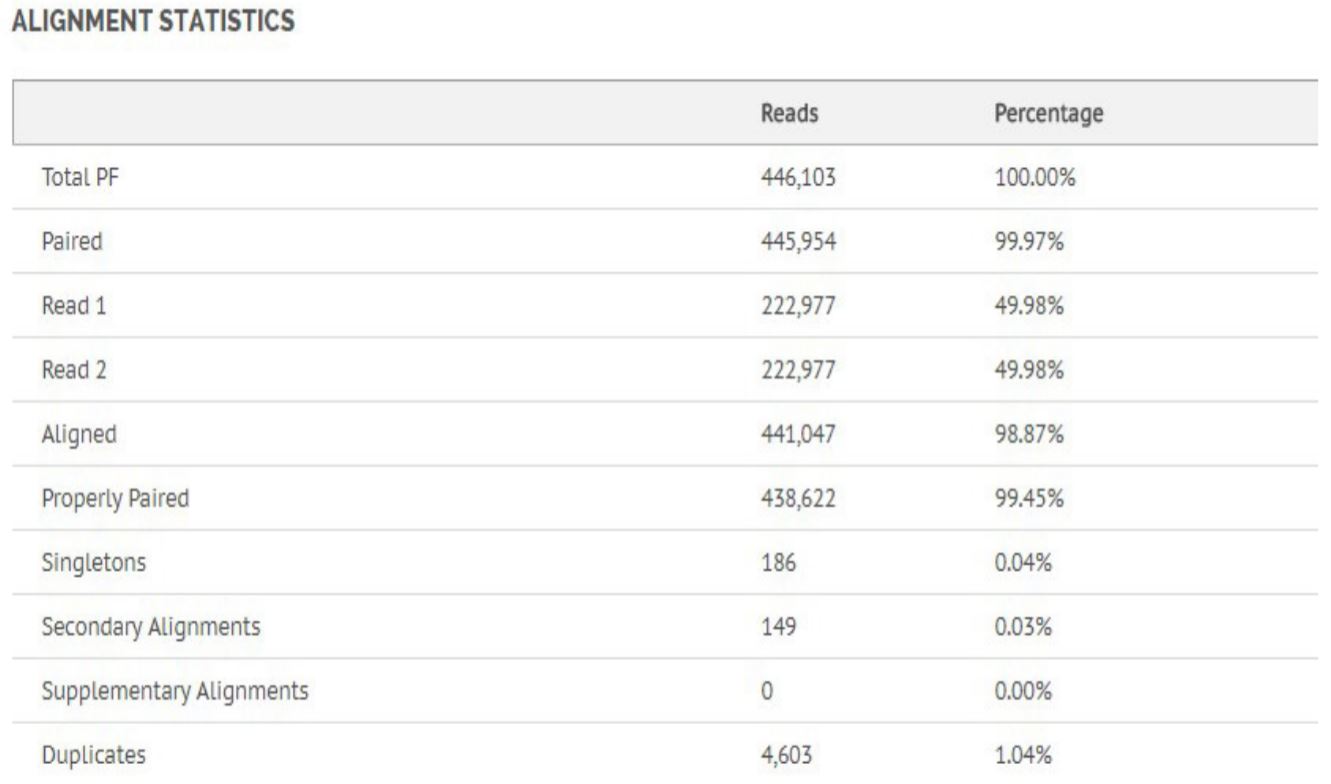
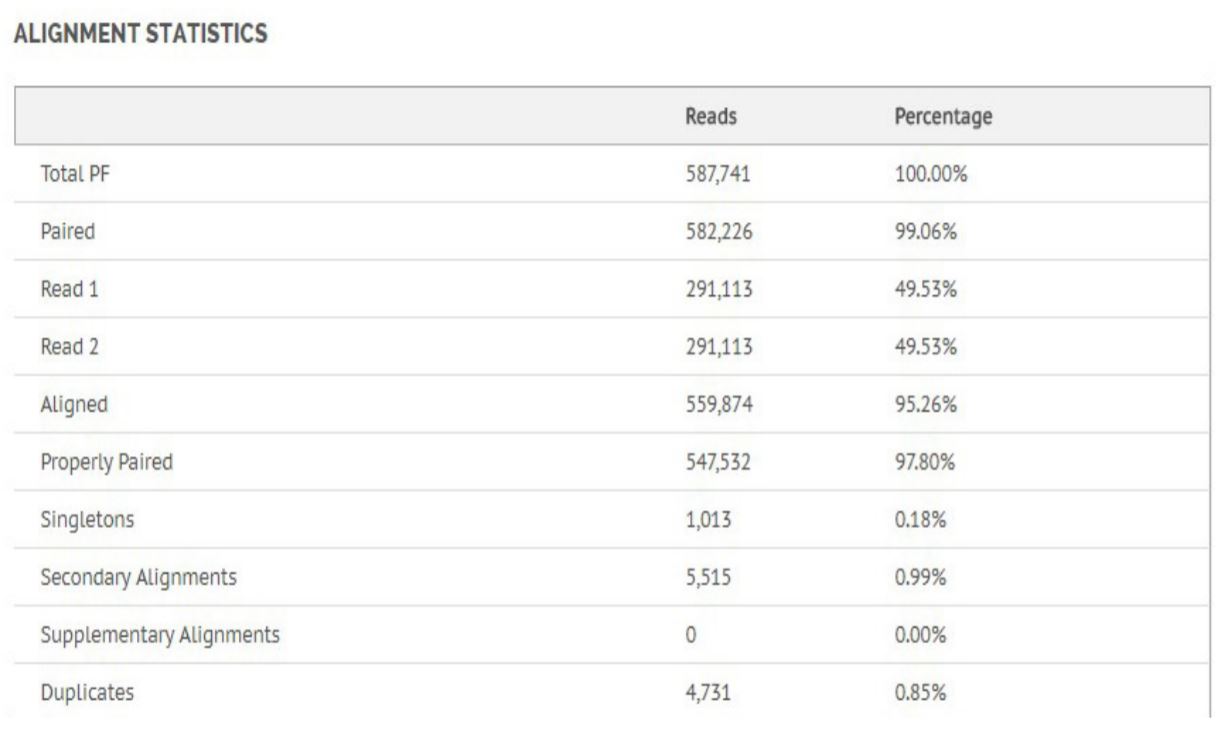
Figure 8. Alignment statistics for Bacillus Cereus.
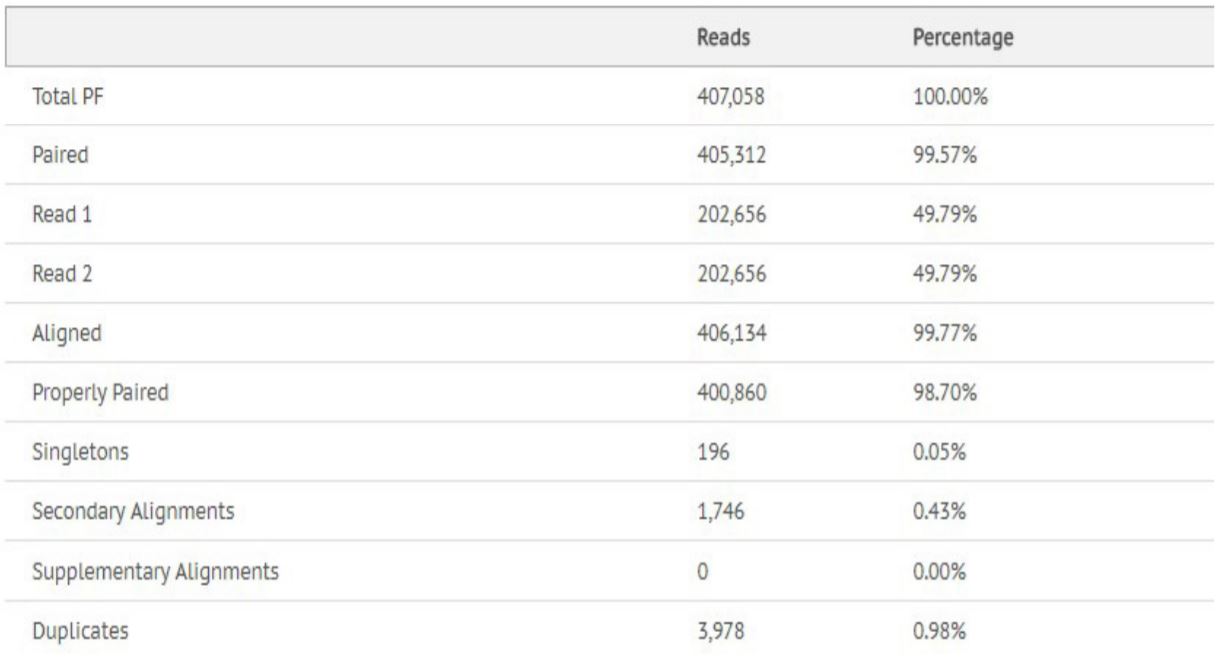
Figure 9. Alignment statistics for Escherichia Coli.
Figure 10. Alignment statistics for Rhodobacter Sphaeroides