Introducing the Cydem VT System: A high-throughput platform for fast and reliable clone screening in CLD
Figure 1. The Cydem VT Automated Cell Culture System.
Introduction
The Cydem VT System (Figure 1) is a high-throughput cultivation platform for automated clone screening and selection. The system combines several technologies to allow for parallel fed-batch cultivation and monitoring of 96 bioreactors with individual gassing. Online, non-invasive pH, dissolved oxygen (DO) and biomass measurements provide continuous control over critical cultivation parameters. On-deck determination of cell concentration, viability and titer concentration ensure selection of the top clones.
In this application note, we describe how to set up your experiment on the Cydem VT System. An overall understanding of the device’s modules will be given, and protocol and experiment considerations discussed. Furthermore, the options for data analysis are presented. A more extensive overview of the system’s components, operation principles and instructions can be found in the Instructions for Use (IFU) manual.
Cydem VT System Breakdown
The Cydem VT platform consists of several technologies to allow for the cultivation and monitoring of Chinese Hamster Ovary (CHO) cells in cell line development. A bioreactor designed for cell culture experiments is found at the heart of the system. It allows for 96 parallel cultivations in shaken microtiter plates with a working volume of up to 8 mL. The bioreactor is flanked by two analytical modules: the Cell Health module and the Titer module. These will deliver crucial answers about the status of the cultures. The modules are placed within a Liquid Handler that will take care of all the liquid handling steps – such as feeding of the cultures and liquid pH adjustments – which allows for extended walkaway times. Beckman Coulter Life Sciences will provide all required reagents and consumables in dedicated experiment kits to ensure smooth experiment setup. In the following sections, a brief breakdown of the four main components and the experiment kits of the Cydem VT System is provided.
Bioreactor Module
The integrated bioreactor module allows for the parallel cultivation of up to 96 clones distributed over four 24-well bioreactor plates. The bioreactor will shake the plates to ensure optimal gas transfer and homogeneous distribution of cells and nutrients. It will also control the supply of N2, CO2 and O2 to all 96 cultivations. Furthermore, the bioreactor module provides important cultivation data using the optical measurement system. Through scattered light detection, an online biomass signal is provided and pH and Dissolved Oxygen (DO) are determined using optodes spotted into the bioreactor wells. These online measurements are used to continuously monitor and control pH and DO values.

Figure 2. Liquid handler interacting with the bioreactor.
The bioreactor plates are clamped into the bioreactor module at the start of the experiment and will remain in their position until the experiment is completed. A self-sealing TPE seal closes the system, and conical guides allow access for adding or withdrawing liquids using a combination of both fixed and disposable tips. Furthermore, the bioreactor plates are equipped with a gassing lid containing four microfluidic gassing channels per cultivation well (Figure 3). Three of these channels are used to provide every well with O2, CO2 and N2, the remaining channel is used as an exhaust gas channel. Adjusting the flowrate of the individual gases allows the bioreactor to change the composition of the overhead gas mixture per bioreactor, e.g., adding additional CO2 results in a lower pH, increasing the amount of O2 will increase the DO value.

Figure 3. A single bioreactor plate, allowing for individual O2, CO2 and N2 gassing through the gassing chip (left). During the cultivation, online measurement of DO and pH provide culture insights. Typically, a Cydem VT experiment will run with four bioreactor plates.
Cell Health Module
The integrated Cell Health Module provides cell viability and density measurements for samples taken from the bioreactor module. All necessary Cell Health reagents are stored in the front right of the instrument deck. The module relies on an automated trypan blue dye exclusion method to determine both cell concentration and viability. Only 43 μL per cultivation is needed, which will allow users to sample their cultivations daily. The module can measure cell concentrations up to 60 million cells per mL. Similar to the Vi-CELL BLU Cell Viability Analyzer, the Cell Health module allows users to customize and optimize the Cell Type parameters used for analysis.

Figure 4. The liquid handler dispenses a sample into the Cell Health Module.
Titer Module
The Titer Module is integrated on the left side of the deck and allows for automated determination of IgG concentrations using a fluorescence polarization based assay. The assay relies on pre-coated IgG-detecting titer plates. The probes inside these plates will be rehydrated with diluent briefly before the sample from the bioreactor is added. The IgG antibody present in the sample will bind to the rehydrated probes in the titer plates, which will result in an altered fluorescence polarization which can be quantified and – using an IgG Standard Curve – used to calculate the samples’ antibody titer. For titer determination, only 10 μL of sample is needed. Running samples on both the Cell Health and the Titer Modules will thus require a total sample volume of only 53 μL.

Figure 5. The liquid handler dispenses a sample into the Titer Module. Right next to the Titer Module there are several Titer Plates (sealed with silver foil), containing the IgG-detecting probe.
Liquid Handling System
The Liquid Handling System takes care of all the liquid transfers from and to the modules and deck positions. The Liquid Handling Pod consists of eight individual probes – evenly split between fixed and disposable tips. Furthermore, a 360 degree rotating gripper allows the system to transfer and stack labware as needed during the experiment setup and run. Below the deck, several containers are stored to supply DI water and decontaminant (like 0.5 M NaOH) for tip washing, and to collect liquid waste generated by tip and module washing steps. Also, a Trash Chute is positioned on the far-left side of the deck where used labware and tips can be discarded. The discarded consumables are collected below the deck and can be removed via the workstation cart.
Figure 6. Liquid handler aspirating a culture additive.
Experiment kit
Beckman Coulter Life Sciences will supply complete experiment kits to ensure swift experiment setup. The kit will contain four bioreactor plates, ten titer plates and all deck consumables like sterile tips, mixing plates, sample plates, reservoir plates and lids. Additionally, the kit comes with fresh Trypan Blue, Cleaner and Diluent for the Cell Health and Titer Modules. The kits provide ample reagents and consumables for a typical, two-week clone screening experiment in 96 parallel reactors.
Protocol and experiment setup
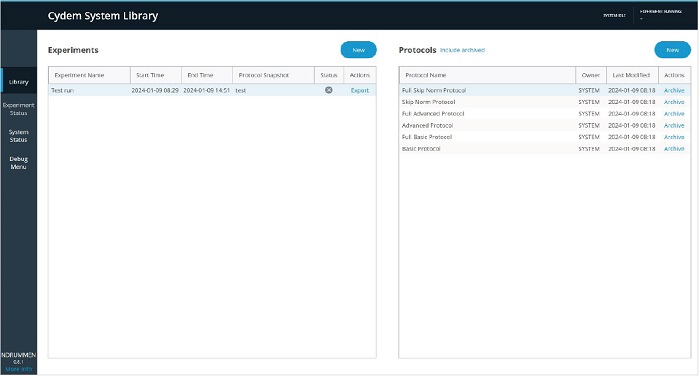
Before starting any experiment, make sure to follow all recommended cleaning steps as described in the IFU manual. Make sure all waste containers are emptied and all reagent containers filled. Follow all recommended quality control procedures to ensure proper functioning of the system. The Cydem VT software will provide guidance during protocol creation, experiment preparation and experiment execution. Protocol creation and experiment setup are found in the Library tab (Figure 7) of the software.
Protocol creation
A protocol defines the cell culture cultivation parameters, and designing a protocol is required prior to configuring and starting a cultivation experiment. One protocol can be used to start multiple consecutive experiments with identical environmental conditions and setup. The protocol configuration wizard will guide users through the protocol creation steps. First, select New in the protocol section of the Cydem System Library and add a Protocol name, description, and duration. Then, define the Starting volume and the Bioreactor Plates used (Figure 8).

Figure 8. Step 2 of protocol design.
Step 3 consists of setting the environmental setpoints temperature, shaking frequency, pH and DO for the cultivation (Figure 9). The pH control depends on three parameters: The Base Target Point, Base Trigger Point and CO2 Setpoint. Typically, cell culture media require CO2 gassing to reach the optimal pH for cell growth. Select the desired pH for the CO2 Setpoint and the Cydem VT System will control the culture’s pH by increasing or decreasing the concentration of CO2 in the headspace of the bioreactor plates. Usually, the amount of CO2 required to keep the pH at setpoint will decrease during the first days of culture due to the acidification of the cultivation as the cells produce lactic acid. When a certain amount of lactic acid has been produced, the pH will drop below the setpoint even without the presence of any CO2 and liquid base addition is required to restore the desired pH. Once the pH drops below the Base Trigger Point, a base addition will be triggered and executed during the next scheduled base addition. The Base Target Point is the (user-defined) desired pH after finishing the base addition step. The Cydem VT software will estimate how much base needs to be added to reach the Base Target Point.
Important: The Base Target Point and the CO2 Setpoint don’t need to be identical. If the culture’s pH reaches a higher value than the CO2 Setpoint after a base addition step, CO2 gassing will be used to bring down the pH to the CO2 Setpoint.

Figure 9. Step 3 of protocol design.
Next, define the reagents added on the deck (Figure 10). Add the Base concentration and scale factor. Add a Name and Liquid Type for all desired Feeds and Additives and check Premix if they are miscible with other scheduled feeds and additives before dispensing into the bioreactor. Premixing will be performed on a mixing plate and will reduce the time the bioreactor cover needs to be opened as it will reduce the number of liquid handling steps from the deck into the bioreactor plates. The feed schedule needs to be defined in step 5 (Figure 10) and allows for distinct schedules per plate, or alternatively, feed strategies can be copied from one plate to another.

Figure 10. Steps 4and 5 of protocol design.
After defining the feed schedule, the desired testing schedule needs to be determined (Figure 11). The Cydem VT System allows for daily Cell Health and Titer measurements, but fewer sampling timepoints can be selected if desired. Scheduling a Titer Test will automatically schedule a Cell Health Test on the same day, but Cell Health Tests can be run without scheduling a Titer Test. Finally, the system will estimate how many kits are needed for running the created protocol and allows the user to set preferred times for testing, feeding and base addition. The software will validate the protocol design and prompt the user to fix any errors within the protocol before saving (Figure 11).

Figure 11. Steps 6 and 7 of protocol design.
Creating a new experiment
Once the protocol has been saved, preparation for a cell culture experiment can be started. Make sure the system is clean before starting the experiment configuration with the Cydem VT Software and decide which seeding strategy is desired and make sure all data from the previous experiments has been retrieved. Also, gather all labware, materials, reagents and kits needed for your experiment setup. Before starting an experiment with Titer Tests, an IgG standard curve needs to be created. The Cydem VT System will support users creating standard curves for their desired standard. Then, select New in the Experiments section of the Cydem System Library (Figure 7).

Figure 12. Starting a new experiment.
Add an Experiment name and description, and select the Protocol and IgG Standard Curve to be used (Figure 12). In the next section, individual bioreactor well names can be assigned to allow for easy identification of the cultures. Scanning the QR code on the consumables kit will auto-populate the relevant details for the bioreactor plates to ensure the correct pH and DO Calibration Data is used.

Figure 13. Insert bioreactor plates lot number and calibration data.
Now, the bioreactor plates can be placed into the bioreactor and clamped down. Make sure the plates are in the correct position and clamped down evenly. The software will guide the user through all steps prior to inoculation of the bioreactor plates. Once all bioreactor plates are seeded, the experiment is started, and the Experiment Status tab (Figure 14) is populated. There, an overview of the running experiment is given including the Reagent Status, Alerts, Bioreactor Module conditions and the Experiment Calendar.

Figure 14. Experiment Status tab.
Running the experiment
The Experiment Status tab is the central page that provides users with the actual status and upcoming actions for the ongoing experiment. The Experiment Calendar provides an overview of the performed and scheduled activities. The Time to Setup Required section shows the user when the next scheduled activity will start, and it will indicate when labware or reagents need to be replaced. Make sure these are replaced on a timely basis so that the system can execute all actions as scheduled.
The Bioreactor Module conditions section provides information for all individual wells. Select one (Figure 15) to get the latest measurements from the optical system (pH, DO, Biomass) and the Cell Health and Titer Module (TCD, VCD, Viability and IgG Concentration). Navigate to the System tab to get detailed information on all consumables and reagents. Select any module to see its status and – when needed – refill or replace reagents. Please refer to the IFU manual for any alerts or alarms that show during an experiment.

Figure 15. Bioreactor Module conditions. The left-bottom corner shows the most recent data of the selected well (Plate 1, B2).
If desired, several Manual Actions can be performed during the experiment, including generating an offline plate for additional sample analysis, adding an additional feed to the schedule, and terminating wells before the experiment is completed. These actions can be performed in between the scheduled actions. Select View Protocol to get more detailed information on the protocol that was used to run the ongoing experiment or export the experiment data to analyze or visualize the results up to the current state.
End experiment
Once the experiment is ended, the system will stop all feeding, liquid pH control and testing activities. Nevertheless, the bioreactor will continue to shake and gas in case the bioreactor wells need to be harvested for post-run analysis or sample storage. If harvesting is required, the Cydem VT software will guide the user through this process to ensure all required labware is available. Afterwards, the bioreactor plates can be removed, and the final experiment result file exported. Now the system can be cleaned, and labware and reagents can be replenished to prepare the system and deck for another run. Please make sure to follow the instructions provided by the IFU manual regarding system tear down, cleaning, decontamination, quality control and maintenance.
Data analysis
Data from a completed or ongoing experiment can be analyzed and visualized using any offline data analysis software. Export the csv file containing all relevant data and conduct the desired data analysis directly at the Cydem VT Workstation or remotely at any desk. An example of experimental data analysis can be found in the next section.
Clone screening experiment
To demonstrate the power of the Cydem VT Automated Cell Culture System, a CHO fed-batch clone screening experiment was conducted with 6 different clones. Per clone, 4 cultivation wells were used to test the reproducibility between bioreactors (Table 1).
| 1 |
2 | 3 | 4 | 5 | 6 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Clone 1 |
Clone 2 | Clone 3 | Clone 4 | Clone 5 | Clone 6 | |||||||||
| B | |||||||||||||||
| C | |||||||||||||||
| D | |||||||||||||||
Table 1. Well map for the clone screening experiment in the Cydem VT System.
The experiment was conducted on the Cydem VT System. For seeding, the unknown VCD workflow was used, with a targeted starting cell density of 0.4*106 cells/mL. The cultivation was started, running at 37° C and 800 rpm. The CO2 Setpoint was set at 7.1, the Base Trigger Point at 6.9, and the Base Target Point at 7.0 – for liquid base adjustments, 0.5 M NaOH was added to the deck.
Online determination of the cell concentration and viability was scheduled daily, and from day 3 onwards the cultivations were supplied with a feed solution and additional glucose. The feed schedule was based on results from previous experiments. Online antibody titer determination was scheduled from day 6 onwards.


Figure 16. Cell concentration and viability during the 14-day fed-batch run, determined by the Cydem VT System. The error bars represent the standard deviation per clone.
The cell concentration and viability data (Figure 17) shows distinct growth profiles for the individual clones. Furthermore, the excellent reproducibility per clone resulted in low coefficients of variation (CV) per clone per day. The average CV across all measurements was 5.4% for the total cell density, and 0.9% for the viability. All selected clones grow over the course of the cultivation experiment, but the maximum viable cell concentrations vary between 11.6*106 and 23.7*106 cells/mL.

Figure 17. Titer value on day 6, measured on the Cydem VT System.
The online titer values (Figure 17) show low variability between the clone replicates, at an average CV of 6.3% for all clones. The tested clones’ titer values at day 14 vary between 1.4 g/L for clone 5 and 4.4 g/L for clone 3. The combination of the available cell concentration and viability, and titer data allows for confident screening and ranking of CHO cell clones with the Cydem VT Automated Clone Screening System.



