Whole genome sequencing is a powerful technique allowing the complete DNA sequence of an organism to be determined, and careful sample preparation is the key to success. An essential step in the sample preparation process is cleanup, which is vital for ensuring the accuracy and reliability of sequencing results. Sudha Savant, Commercial Product Manager for Genomics at Beckman Coulter Life Sciences, highlights the significance of sample cleanup in sequencing, and how we can help.
The critical role of nucleic acid cleanup: ensuring sequencing success with quality reagents
Whole genome sequencing has come a long way since its emergence in the 1970s.1 Decoding an organism’s genetic material was originally a laborious and expensive process, but sequencing has now transformed into a rapid, accurate, and widely available technique that offers crucial insights into biology and medicine.2 To prepare samples for next-generation sequencing (NGS), several key steps must be carried out, including nucleic acid extraction and library preparation. In addition, it is necessary to remove any impurities that could interfere with sequencing reactions or compromise the quality of results.

Figure 1. Example of NGS workflow
The targeted removal of unwanted fragments and contaminants – including enzymes, NGS adapters, primers, primer dimers, unincorporated dNTPs, and other reaction components – is referred to as sample cleanup. This step is often overlooked in sequencing workflows, but it is essential for achieving precise, reliable, and reproducible results. Neglecting cleanup can lead to compromised sequencing data, reduced confidence in outcomes, and the need to repeat experiments, increasing both cost and time investments.
Common nucleic acid cleanup challenges
Effective cleanup is an essential part of the sequencing process, but purifying samples comes with several common challenges.3
Low recovery rate
The first obstacle standing in the way of efficient library purification is the risk of sample loss. A clean DNA or RNA sequence is often a prerequisite for genomic applications like NGS and PCR, so cleanup must ensure that any unwanted material is removed without losing valuable genetic content, particularly low copy number sequences. Although some sample loss is unavoidable, inefficient cleanup can greatly decrease the genetic information available for sequencing, limiting the insights that can be gained.
Cost and time implications
Poor NGS cleanup or PCR cleanup can result in sequencing failures or low-quality data, which may make it necessary to repeat genomic workflows. This results in increased overall time and cost of sequencing. Sequencing errors are particularly wasteful when dealing with rare cell populations or specimens. Investing in high-quality cleanup kits and reagents can prevent these issues, making the overall process more streamlined and cost effective.
Processing errors
The potential for processing errors also presents an important issue in nucleic acid cleanup. While many low-throughput laboratories perform cleanup steps manually, it is often possible to automate these processes. Automation offers several benefits over manual workflows, as it can reduce user errors and improve precision in NGS data. The choice between manual and automated cleanup methods depends on several factors, primarily sample throughput and budget.
Nucleic acid cleanup strategies
Choosing the appropriate cleanup method is essential to address these challenges effectively. Selection will also depend on factors such as sample volume, purity requirements, available equipment, and the specific sample type. While traditional methods like ethanol precipitation can be labor intensive and time consuming – as well as prone to variability and human error – newer column- and bead-based approaches offer greater efficiency and reliability, making them ideal for modern genomic studies with large sample sizes.
Column-based cleanup
Column-based nucleic acid purification involves passing the sample through a specialized column containing a porous matrix. This matrix selectively binds DNA or RNA and, after other contaminants are washed away, the purified nucleic acid is eluted from the column. Column-based methods typically yield highly pure samples suitable for downstream sequencing applications, and kits are available in various sizes to accommodate different sample volumes. However, this process can be time-consuming and costly, involving multiple steps and expensive equipment, such as centrifuges.
Bead-based cleanup
During bead-based cleanup, magnetic beads that selectively bind nucleic acids by type and size are added to the nucleic acid sample. The buffer used in this process creates salt bridges, causing the nucleic acids to bind to the beads. After binding, the beads are separated from the sample using a magnetic field, the rest of the sample is discarded, then the bound nucleic acid is washed and eluted. Bead-based methods are often faster than column-based techniques, as they involve fewer steps, do not require centrifugation, and can be more easily automated. This makes them particularly well-suited to high-throughput applications.
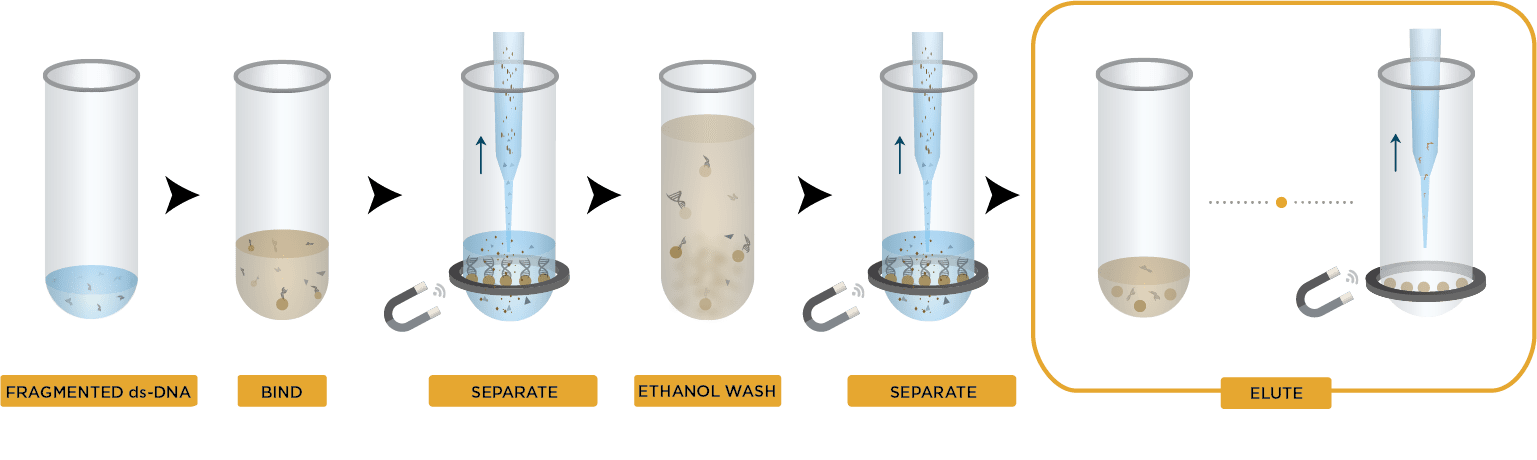
Figure 2. Bead-based nucleic acid cleanup workflow
The importance of investing in premium cleanup reagents
Finally, investing in a high-quality DNA or RNA cleanup kit is possibly the most important decision when it comes to generating reliable sequencing results. Although these solutions may have a higher upfront cost, they offer superior performance and efficiency compared to lower quality alternatives – including ethanol precipitation – minimizing the risk of incomplete contaminant removal or nucleic acid degradation to prevent unreliable data or poor results. Ultimately, this investment saves time and resources by reducing the need for troubleshooting or repeating experiments.
We offer a growing portfolio of genomic solutions – including nucleic acid purification and cleanup reagents – designed to accommodate a range of input materials, as well as automated and semi-automated workflows. Our reagents are designed to effectively remove contaminants, while preserving the integrity of nucleic acids and maximizing their recovery. Our high-performance SPRI bead technology uses magnetic beads to selectively immobilize nucleic acids by type and size, using optimized binding conditions to enable highly specific separation and cleanup protocols. This ensures consistent, reproducible results for higher confidence in sequencing outcomes.
Contact us to find out more about how we can support your cleanup protocols and help you to obtain accurate and reliable DNA or RNA sequencing results.
References
- https://www.nature.com/articles/d42859-020-00100-w
- https://www.yourgenome.org/theme/timeline-the-past-present-and-future-of-sequencing-technologies/
- https://www.cytivalifesciences.com/en/us/news-center/next-generation-sequencing-main-challenges-and-solutions-10001
About the Author

Sudha Savant, a commercial product manager for Genomics at Beckman Coulter Life Sciences, has a background in biochemistry and molecular biology. She has experience with automation of genomic extraction and sequencing solutions and proteomics for applications in areas of oncology, epigenetics, and small molecule discovery. Currently, Sudha manages the bead-based cleanup product portfolio at Beckman, which harnesses the SPRI technology. Her goal is to collaborate with internal and external researchers to develop new applications for the cleanup products.
Featured Products
Not intended or validated for use in the diagnosis of disease or other conditions.
© Beckman Coulter, Inc. All rights reserved. All rights reserved. Beckman Coulter, the Stylized Logo, and Beckman Coulter product and service marks mentioned herein, including Biomek, are trademarks or registered trademarks of Beckman Coulter, Inc. in the United States and other countries. ECHO is a trademark or registered trademark of Labcyte Inc. in the United States and other countries. Labcyte is a Beckman Coulter company. All other trademarks are the property of their respective owners.