Automating Cell Line Development
Summary
- Cell line development is a multifaceted process that stretches over months.
- Automation on a Biomek i7 Workstation helped to overcome many challenges for this workflow including:
- Sample contamination
- Consistent pipetting of semi-solid media
- Monoclonal cell plating by limiting dilution
- Hit picking for IgG analysis and cell expansion
- Integrating devices can enable:
- Reduced hands-on time moving labware
- Cell counting for in-process calculations of transfer volumes
- Centrifugation of cell plates for immediate monoclonality determination
- Automated confluence and titer measurements to drive hit picking
- Sample and data integrity across multiple analyzers over months
Introduction
Cell line development is the process by which the cellular machinery is co-opted to manufacture therapeutic biologics or other proteins of interest. This most frequently involves the production of antibodies in Chinese hamster ovary (CHO) cells grown in suspension cultures for eventual use in bioreactors at the manufacturing stage. The development of these cell lines is a long and arduous process with numerous steps and challenges (Figure 1).
Figure 1. Cell line development process. Automated steps described here are indicated in red.
Once CHO cells have been transfected with the construct of interest a selection step is required to enrich for those cells that have taken up the construct. Single cells must then be isolated, grown into colonies, and assayed for protein production. Once this production screen has been accomplished, the top candidates are expanded and maintained to test for expression stability. Eventually the top hits are further optimized in different media conditions to enhance expression or other protein attributes (e.g. glycosylation) before scaling up to production levels.
Because this process takes months to complete, there are many more challenges than a typical short-term cellular workflow. First, maintaining sterility is essential during various manipulations and throughout the weeks of growth from a single cell. Secondly, as with most screens, the throughput required to identify a handful of optimal clones typically requires thousands of wells. This throughput then tapers off after the hit picking phase, so one should schedule overlapping campaigns to maximize resources. Finally, as clones move through the system and are reformatted into lower density plates during hit picking, all previously recorded data must be tracked as these cell lines are established. We have overcome these challenges by automating the establishment and screening of clones for cell line development.
Automated cell line development system
All cell manipulations and sample preparation was performed on a Biomek i7 Workstation with Span-8 pipettors and a 1200 μL capacity 96-channel head (Figure 2). A HEPA-filtered enclosure and sterile pipette tips were utilized throughout the experiment. The Biomek was integrated with a Cytomat 2C incubator (Thermo Fisher), a Microplate Centrifuge (Agilent), and a Vi-CELL BLU Cell Viability Analyzer. Cell imaging on a CloneSelect Imager (Molecular Devices) and protein quantification on an Octet HTX (Pall ForteBio) were performed offline but both devices can be integrated with the Biomek i7 Workstation – either directly or with the use of a robotic transport.

Figure 2. Biomek i7 Workstation with various integrated devices.
Selection and enrichment
The work described here utilized the DP12 line of CHO cells expressing IgG molecules against interleukin 8 (ATCC). In the absence of selection agents, we found the cells had variable expression that well represented the establishment of stable expression lines.
To enrich the population of cells secreting IgG prior to clonal expansion, CHO cells were counted on an integrated Vi- CELL BLU and 18,000 cells were dispensed to 22 mL semi-solid CloneMedia CHO Growth A media containing a 1:100 dilution of CloneDetect reagent (Molecular Devices). Using fine pipetting control, cells were slowly resuspended in the media and dispensed to 6-well plates without introducing bubbles (A). Plates were incubated for 12 days to form colonies and then analyzed and picked using the ClonePix 2 (Molecular Devices, Figure 3A)
Figure 3B shows an image of the 6-well plates taken in brightfield and FITC channels with the ClonePix 2. Colonies that were secreting IgG showed a halo of fluorescence as IgG molecules aggregated near the colony within the semi-solid media and were stained by the CloneDetect reagent. Based on characteristics such as colony size, shape, and external FITC intensity, a subset of colonies were picked (Figure 3C) into 96-well plates containing 200 μL of XP CHO Growth A media for further culture, analysis, and monoclonal colony formation.

Figure 3. Enrichment of secreting cells. The ClonePix 2 (A) uses imaging to identify clones in semi-solid media based on size (cell growth) and external fluorescence (antibody secretion, B). C) Optimal clones are then picked from the semi-solid media and transferred to 96- well plates for suspension culture.
Isolate single cells and expand
To initiate single cell colonies, suspension CHO cells were mixed and dispensed to an integrated Vi-CELL BLU Cell Viability Analyzer for counting. ~83,300 cells were then stained with 0.25 μM calcein AM at 37°C for 30 minutes. Following an initial 25-fold dilution, ~467 cells were added to 140 mL XP CHO Growth A media (Molecular Devices) such that there was one cell per 300 μL on average. Robust mixing and rapid plate filling was achieved through the ability to aspirate and dispense 96mL at one time with the 1200 μL-capacity multichannel head. Plates were spun in the integrated microplate centrifuge at 300 X g for 3 minutes to ensure all cells were at the plate bottom for immediate imaging on the CloneSelect Imager (Figure 4A).
After plating, wells were imaged in brightfield and fluorescent channels to count calcein-stained cells. As expected with a limiting dilution approach, roughly 33% of wells had a single cell across five experiments (representative plate shown in Figure 4B). Cells were allowed to grow for three weeks and cell confluence was assessed at multiple time points. By using conditioned media (see Discussion) we achieved growth in roughly 50% of the monoclonal wells, resulting in ~15% of the plated wells being eligible for establishment of a permanent cell line (Figure 4C, pink highlights).
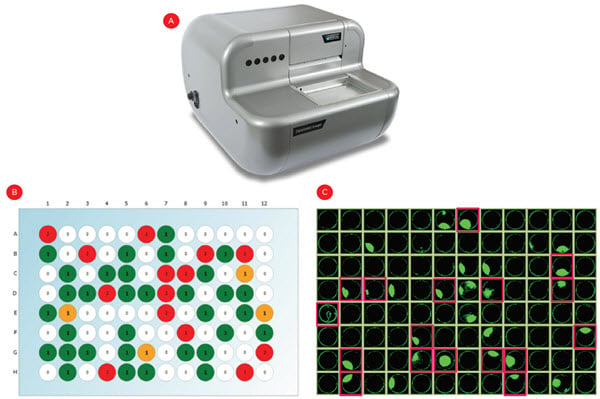
Figure 4. Identifying single cell-derived colonies. Following limiting dilution cell plating, the CloneSelect imager (A) was used to count fluorescently-stained cells in each well to identify monoclonal wells (B, green wells). (C) Cell confluence was also measured over three weeks to identify the monoclonal wells that grew into colonies and warrant testing for IgG expression (pink highlights).
Assay protein production
In a typical screen, all wells would be assayed for the relevant metric. However, since only 15% of the wells have the potential to be taken forward, we chose to sample media only from the wells with colony growth from a single cell. Using the Span-8 pipettors, media was transferred to replicate wells of a 384-well Tilted Well Plate for analysis on the Octet HTX (Figure 5A) and protein A-coated biosensors were used for IgG quantification.
For IgG analysis, binding curves were generated with known Protein A standards (Figure 5B) and compared to binding curves from the assayed wells (Figure 5C) to establish IgG concentration (Figure 5D). Wells with high expression were then resuspended and transferred to larger-area 24-well plates for further expansion and downstream analysis.

Figure 5. IgG quantification. The Octet HTX (A) was used to measure binding curves of known standards (B), and wells with single-cell derived colonies (C). The standards were used to quantify the IgG from the unknown wells and the highest producing clones (D, circled) were moved forward in the process.
Integrated cell line development system
The devices described here can be integrated into a single workstation that reduces human intervention and thereby saves time and lowers the opportunity for errors (Figure 6). In addition, data from the various analyzers can be directly utilized to drive downstream steps (e.g. hit picking), without the need for manual data transfers or manipulations. The suite of SAMI software can manage the processing of multiple plates through the integrated system and track where cells are within a multi-week application such as cell line development. In addition, DART software acts as a data repository that stores data throughout a process or between processes and ensures that the data is easily accessible for additional analysis. By integrating the devices, both sample and data integrity are improved throughout the lengthy workflow.

Figure 6. An example of an integrated cell line development system (front and back).
Discussion
Here we have described an automated solution that overcomes the most complex challenges of cell line development. We were able to maintain cells in a sterile state through multiple manipulations over a 5+ week time course. The Biomek i7 Workstation is able to automate these manipulations at the throughputs necessary for cell line development and the CloneSelect Imager and Octet HTX are able to match this throughput for clone assessments. Integrating these devices further reduces the effort necessary to execute this workflow and improves data traceability by removing the opportunities for errors when handling labware or data files.
Despite these accomplishments, additional biological challenges needed to be overcome. First, while the limiting dilution step was successfully automated, we initially found poor cell survival and growth. By first conditioning the culture media with 1X105 CHO cells/mL for two days, we were able to achieve the 50% survival reported above, indicating the poor growth was due to challenges of monoclonal culture rather than poor automated plating. Cell survival was also negatively influenced by higher concentrations of calcein. The necessarily low level of calcein likely contributed to a relatively short period where the stain was visible by imaging, although this is also likely dependent on the specific CHO cell line. This detection time of ~2 hours could potentially limit the plate throughput based on the imager read time (~15 min/plate on the CloneSelect Imager), however staggered times of stain addition and plating could be included in the automated methods to overcome this limit.
Additional confounding factors can be seen at the time of colony screening. While imaging cell confluence is sufficient to determine if cells have grown, the location of the initiating cell within the well (e.g. center vs. edge) can impact the colony shape, such that there is not a direct correlation between confluence and cell number. While it would be desirable to determine IgG output per cell, this is better left to a later stage during cell expansion when a more accurate cell count can be achieved. Another challenge with IgG quantification is the potential for evaporation over three weeks of culture. Specialized labware with evaporation wells can help with this, but for ease of use, we added 300 μL/well of medium to reduce the impact of evaporation compared to lower well volumes. As with cell counts, IgG production can be more accurately determined during cell expansion and genomic stability testing.
While the steps of the cell line development workflow are fairly universal, there are numerous different ways these steps can be performed. Single cell printing or cell sorting could be used as a more efficient means of establishing monoclonal wells. ELISAs or other approaches could be used to titer antibody production, as the automated system could be used to schedule the efficient processing of the necessary sample preparation throughput.
Even the order of steps we describe here might be altered. We utilized imaging to identify the wells that grew from a single cell so that Octet biosensors could be conserved by only assaying the relevant wells. If one lacks the fluorescent imaging capability for monoclonality determination, one could image all the wells in brightfield at the time of plating and assay all wells for protein production. Only those wells with high expression would then be checked visually for a single cell in the initial images.
Finally, additional steps of the cell line development workflow beyond those described here can also benefit from automation. We have automated the basic tasks of cell culture and can thereby relieve the burden of the repeated passaging required to determine clone stability (Consistent Cell Maintenance and Plating through Automation AAG-2585APP04.17. In addition, we have automated a design of experiment approach to optimizing media components for further enhancement of protein production (Automated Optimization of IgG Production in CHO Cells AAG-2148APP11.16). More information on the automation of the enrichment, limiting dilution, and hit picking steps, as well as data handling is available as well (Cell Line Development Data Handling AAG-3996APP08.18, Cell Line Development Hit Picking AAG-3998APP08.18, Cell Line Development Limiting Dilution AAG-3999APP08.18, Cell Line Development Selection and Enrichment AAG-4000APP08.18). By automating as much of the cell line development process as possible, the effort for each campaign will be minimized while the sample and data integrity will be maximized.

