Biomek i-Series Automated Illumina TruSeq® Stranded mRNA Sample Preparation Kit Protocol
Biomek Automated Genomic Sample Prep Accelerates ResearchIntroduction
Illumina TruSeq® Stranded mRNA Sample Preparation Kit protocol converts mRNA from total RNA into a template sample of known origin. The protocol first uses poly-T oligo attached magnetic beads to isolate mRNA. Strand specificity is attained by replacing dTTP with dUTP in the second strand cDNA synthesis step. Knowing the strand origin improves transcript annotation accuracy and alignment efficiency, reducing sequencing costs per sample. Uniform high coverage provided by the Stranded mRNA Sample Preparation Kit protocol ensures its suitability for multiple applications such as discovering alternative transcripts, gene fusions, and allele-specific expression (Illumina TruSeq® Stranded mRNA Library Prep Guide15031047-e). In this technical note, we present the automation of the Stranded mRNA Sample Preparation Kit protocol on Biomek i7 Dual Hybrid (Multichannel 96, Span-8) Genomics Workstation.
When compared to manual operations, the Illumina TruSeq Stranded mRNA Sample Preparation Kit automated on Biomek platform provides:
Illumina approved stop points and method start/stop points
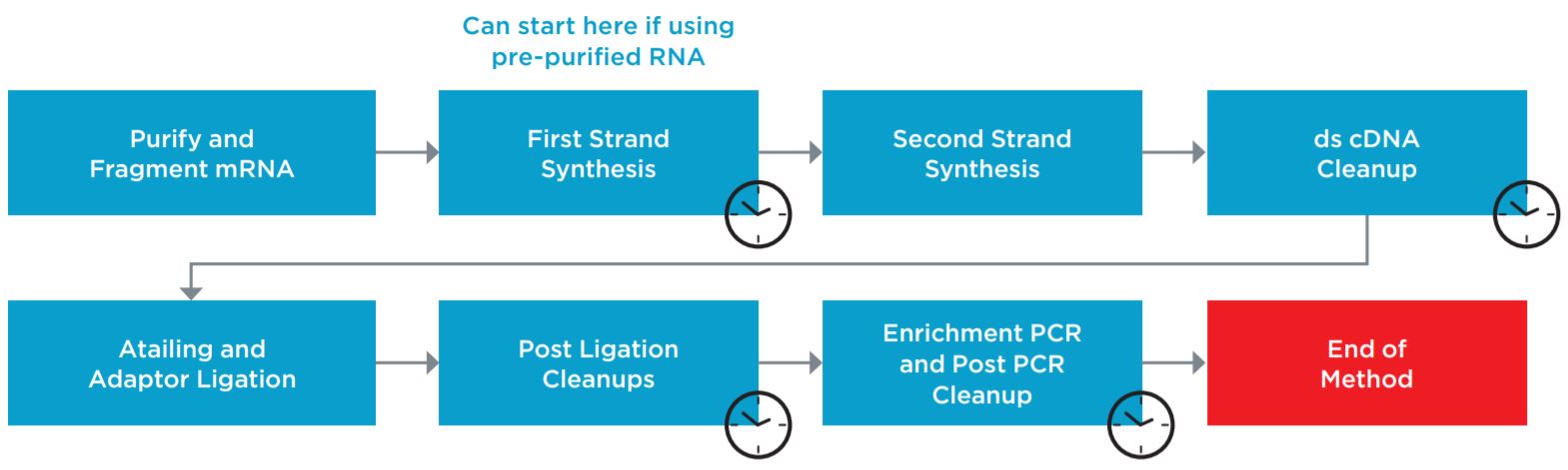
Figure 1. Illumina TruSeq® Stranded mRNA Sample Preparation Kit protocol.
| Process | Time | |
| 24 Samples | 96 Samples | |
| Prepare Reagents, Set up Instrument | 15 min | 30 min |
| cDNA Synthesis | 4 hr, 3 min | 4 hr, 22 min |
| Sample Construction | 3 hr, 13 min | 4 hr, 5 min |
| Total* | 7 hr, 30 min | 8 hr, 57 min |
*Timing estimate includes incubations and thermocycling.
Timing estimate do not include reagent thawing.
Table 1. Estimated run times for Illumina TruSeq® Stranded mRNA Sample Preparation Kit protocol on the Biomek i7 Dual Hybrid Genomics Workstation.
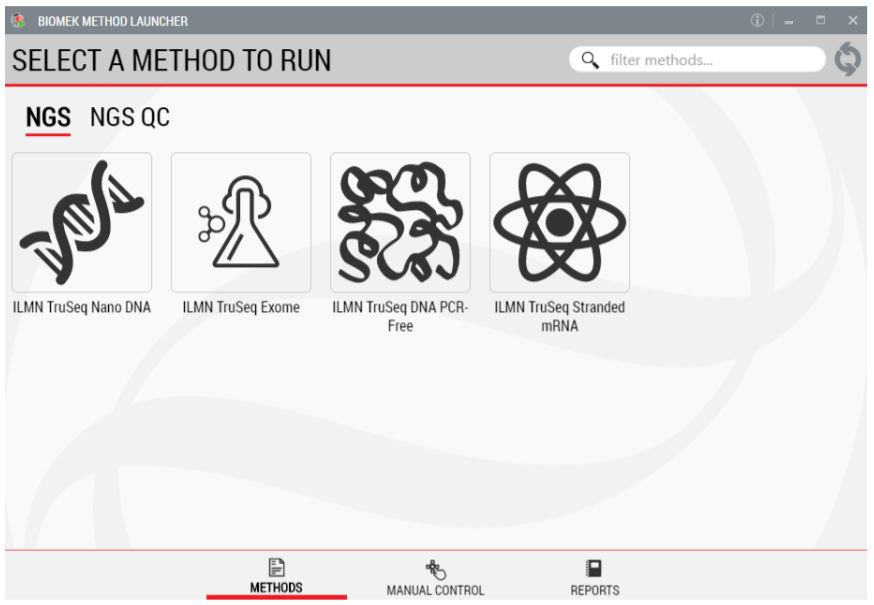
Figure 2. Biomek Method Launcher provides an easy interface to start the method.
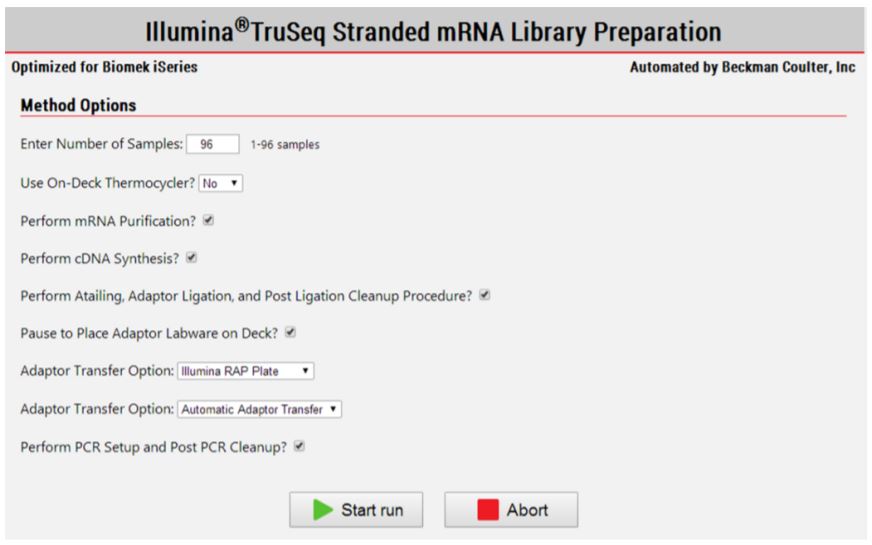
Figure 3. Biomek Method Options Selector indicate sample number and processing options.
Automated Method
Automation of the Stranded mRNA Sample Preparation Kit protocol provides efficient library preparation with minimal hands-on time (Table 1). The automated method is designed to minimize the exposure to hazardous reagents (e.g. Actinomycin D) and to separately collect the reagents for convenient disposal as biohazardous waste. By the use of Biomek Method Launcher, the users can easily implement and adapt the method to suit their needs.
1. Biomek Method Launcher (BML): User friendly interface
BML is a user friendly interface for securely launching the method without introducing errors during method setup (Figure 2). BML enables users to monitor the progress of the run, off site. The method steps are organized in a modular manner for workflow optimization (Figure 1).
2. Method Options Selector (MOS):
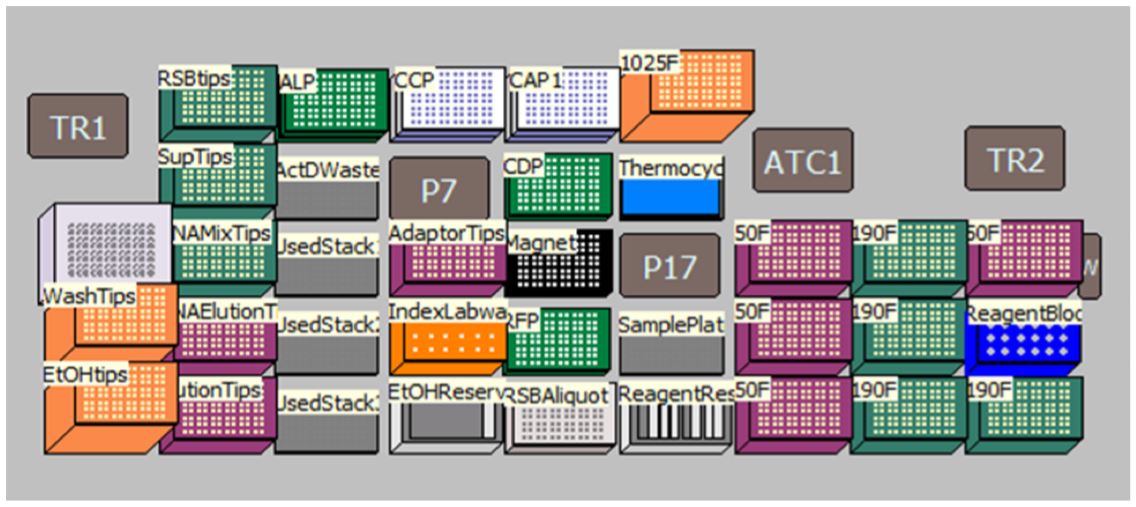
Superior flexibility MOS provides several sample number and sample processing options (e.g. insert size selection, adaptor transfer options, thermocycling options) to maximize flexibility and the adaptability of the method. Modular workflow provides workflow optimization. Ability to start parts of the method based on the user constraints and logical start and stop points assigned based on Illumina’s recommendations, maximizes the adaptability of the method (Figures 1, 3). In addition, optional pause steps are included for placing adaptors on deck. The thermocycling steps can be done either off-deck or on-deck using an automated thermocycler (ATC Thermo Fisher; Figure 4).
Figure 4. Deck Layout for TruSeq Stranded mRNA Sample Preparation Kit protocol on Biomek i7 Dual Hybrid for 96 samples with on-deck thermocycling option. Thermo Scientific’s ATC
3.Guided Labware Setup (GLS): Step by step instructions
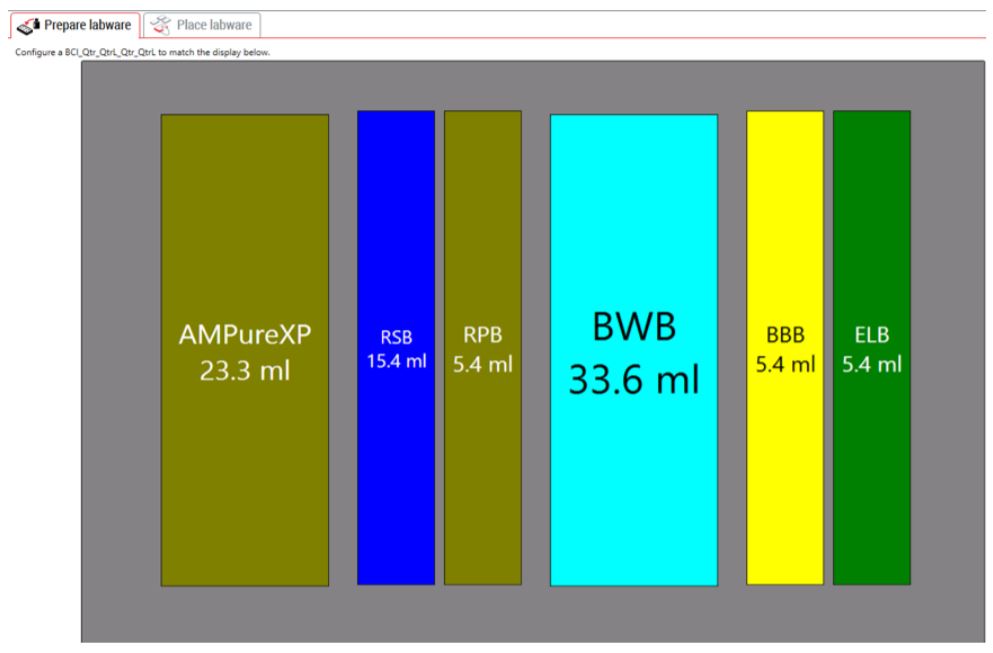
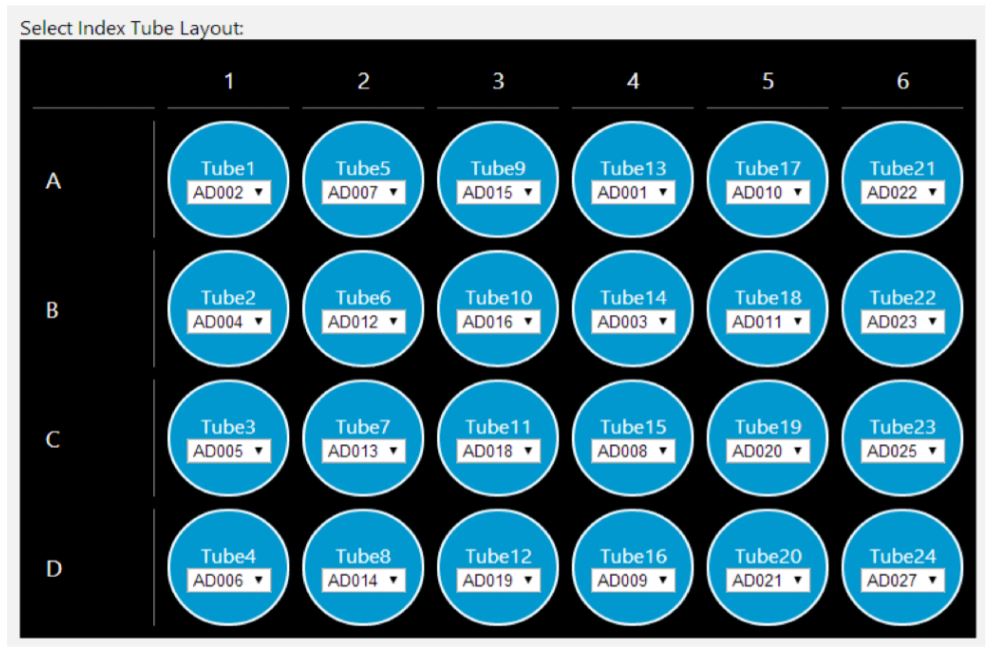
GLS provides the user specific text and graphical setup instructions such as reagent volume calculations and step-by-step instructions to prepare reagents and place labware (Figure 5). The GLS guides the users to place Illumina low throughput and high throughput adaptor labware on deck along with custom adaptor plates (Figure 6). The steps are generated based on the options selected in MOS. For instance, selecting automatic adaptor transfer creates dataset driven adaptor ID logs, indicating which adaptor has been assigned to which sample, through Biomek software Data Acquisition and Reporting Tool (DART). DART gathers data and synthesizes runtime information from Biomek log files to capture all sample manipulations during the course of the method. Alternatively, users also have the option to customize adaptor assignments by uploading a .csv file.
|
Figure 5. Guided Labware Setup indicates reagent volumes and guides the user for correct deck setup |
Figure 6. Guided Labware Setup enables selecting index tube layout |
Experimental Design
Universal human reference mRNA (UHR 500 ng/µl, 250 ng/µl, 50 ng/µl , 25 ng/µl; 4-6 replicates from each)was used for the automation of the Stranded mRNA Sample Preparation Kit protocol on the Biomek i7 Dual Hybrid (Multichannel 96, Span-8) Genomics Workstation with on-deck integrated Thermo Fisher Scientific Automated Thermo Cycler (ATC). After the preparation, the libraries were analyzed on Agilent TapeStation 2200 with Agilent High Sensitivity D5000 ScreenTape system and by qPCR (Applied Biosystems 7900HT Fast real time PCR system) using KAPA Sample Quantification Kit.
Results
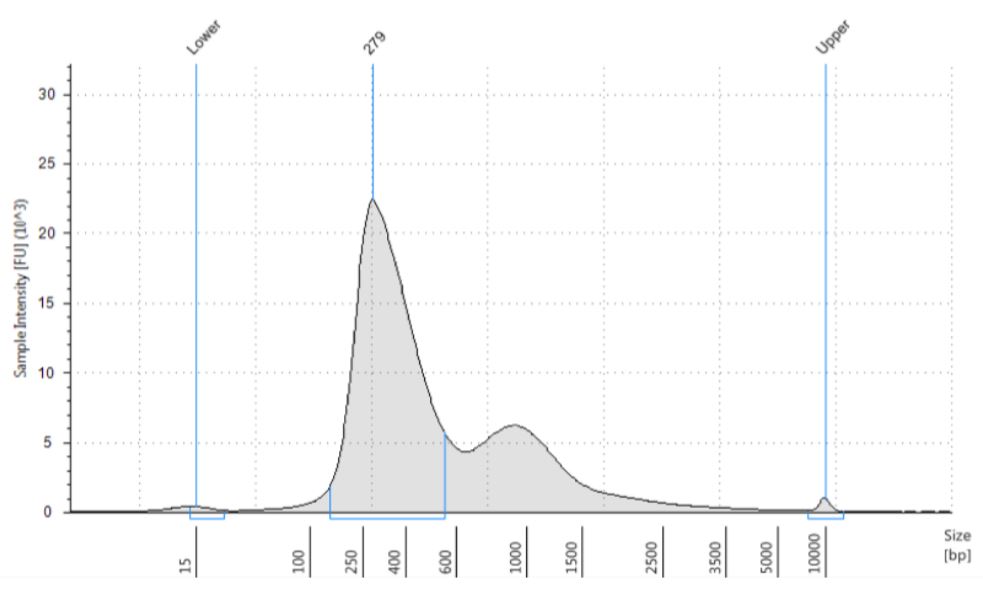
Agilent TapeStation results indicated that the prepared libraries are of expected size for Illumina stranded mRNA libraries (Approximately around 260 base pairs, Illumina TruSeq® Stranded mRNA Library Prep Guide15031047-e; Table 2; Figure 7). As indicated by Illumina, the library yield increased with the amount of input RNA (Table 2). Overall, the quantity and the quality of mRNA libraries are appropriate for the sequencing and downstream applications (Approximately 10nM of samples are used for the proceeding steps, Illumina TruSeq® Stranded mRNA Library Prep Guide15031047-e).
| Sample ID | Index Primer | TapeStation size (bp) | qPCR Yield (nM) |
| UHR 500 Rep3 | AD015 | 293 | 469.3 |
| UHR 500 Rep4 | AD016 | 303 | 443.4 |
| UHR 500 Rep5 | AD007 | 294 | 441.9 |
| UHR 500 Rep6 | AD012 | 301 | 440.2 |
| UHR 250 Rep3 | AD018 | 279 | 299.5 |
| UHR 250 Rep4 | AD019 | 298 | 311.3 |
| UHR 250 Rep5 | AD013 | 283 | 262.2 |
| UHR 250 Rep6 | AD014 | 282 | 305.8 |
| UHR 50 Rep3 | AD002 | 259 | 61.5 |
| UHR 50 Rep4 | AD004 | 256 | 59.5 |
| UHR 50 Rep5 | AD015 | 259 | 50 |
| UHR 50 Rep6 | AD016 | 250 | 51.4 |
| UHR 25 Rep3 | AD005 | 242 | 12.2 |
| UHR 25 Rep4 | AD018 | 255 | 13.6 |
| NTC | AD006 | N/A | 5.2 |
| NTC | AD019 | N/A | 10.6 |
Table 2. Sample quantification of automated Illumina TruSeq® Stranded mRNA Sample Preparation Kit protocol using Agilent TapeStation 2200 and qPCR. NTC: No Template Control.
Figure 7. Electropherogram (Sample intensity vs. size in base pairs) of Agilent TapeStation corresponding to UHR250 replicate3 showing the libraries around expected size of the marker
Summary
We automated the Illumina TruSeq® Stranded mRNA Sample Preparation Kit on Biomek i7 Dual Hybrid (Multichannel 96, Span-8) Genomics Workstation. Our assessments indicate that the quality and the quantity of prepared libraries are in good condition for sequencing. When compared to manual operation, the automated protocol increases the sample preparation efficiency by reducing the hands-on time. In addition, Biomek Method Launcher provides a user friendly interface where the user can easily customize and run the method.