Nanoliter Scale DNA Assembly Utilizing the NEBuilder HiFi Cloning Kit with the Echo 525 Liquid Handler
Jared Bailey, Jefferson Lai, Rabia Khan, and John LesnickAbstract
Previous work has shown the ability of the Echo 550 Liquid Handler to generate two-piece assemblies using the Golden Gate and Gibson Assembly chemistries utilizing a miniaturized protocol.1 In this experiment, NEBuilder HiFi was combined with the 25 nL increment dispensing Echo 525 Liquid Handler. Two unique assemblies were constructed to demonstrate the benefits of modular synthetic biology by rational design. The first assembly contained two pieces. This construct was designed to express the LacZ pathway in a 5223 bp plasmid and allow bluewhite screening when transformed into a non-alpha complementing strain. A second, five-piece assembly with 8,940 bp was also constructed to more closely represent the complexity and size of typical industrial DNA production. A two-piece assembly was successfully miniaturized to 100 nL from a 20 μL recommended volume for a reagent savings of two hundred-fold. For the more complex assembly, reactions were successful down to eighty-fold with forty-fold reductions producing the most robust results. At the forty-fold reduced volume, 1.25 fmoles of DNA was all that was required for each piece in a total reaction volume of 500 nL. The goals of these experiments were to utilize the Echo 525 Liquid Handler in assembling both a two-piece and five-piece assembly in miniaturized reaction volumes. The experiments were validated by junction qPCR for rapid analysis of successful assembly and Sanger sequencing verification for nucleotide resolution. The Echo Liquid Handler enables lower-cost methods and workflows to produce high-quality synthetic DNA constructs which expands design-based testing with higher throughput and affords the scientist a broader biological landscape to interrogate. Ultimately, the utilization of the Echo Liquid Handler for synthetic biology can help close the DNA read-write cost gap.
Introduction
Synthetic biology is an interdisciplinary science with the potential to impact both academic and industrial applications including the creation of novel therapeutics and vaccines, plant science and biofuels, as well as bio-based chemical manufacturing capabilities that involve the application of engineering principles to biology. The focus is often on generating parts of natural biological systems, characterizing and isolating them, and then using them as components of an engineered biological system. A trademark of synthetic biology is the application of rational design principles to the design and assembly of biological components. However, the biology of a synthetic DNA construct introduced into a cell is difficult to predict, it is often necessary to try many versions of the construct to determine which permutation will work best. Therefore, a greater emphasis on the modular design of DNA parts enables the assembly of a greater variety of potential constructs through combinatorial assembly of synthetic components. In addition to simplifying the overall DNA construction workflow, modular design and assembly of DNA components makes automation of the assembly process possible reducing the time, labor, and cost of generating multiple constructs to allow for an increase of throughput with an overall shortened development cycle.
Synthetic DNA plasmids are designed using computer-aided software with experimentally dependent functionality. This can range from interrogation of different domains of a single protein to the production of an entire pathway with heterologous genes. The designed DNA is then divided into synthesizable pieces. The pieces are designed with overlapping sequences to their neighbors before being chemically synthesized. The DNA fragments are then assembled together to build the designed construct using gene assembly techniques. If needed, multiple pieces can be assembled together into larger DNA assemblies. The assembled constructs are then typically cloned into an expression vector and sequence-verified. Once verified, the synthetic constructs are transferred into a production cell and the function of the designer construct is assayed. Depending on the results the constructs can then be modified or refined, and the test cycle is repeated until a DNA construct is obtained that produces the desired function.
New England Biolabs has developed the NEBuilder HiFi DNA Assembly Cloning Kit (Figure 1). The NEBuilder HiFi kit takes input DNA with 15 to 30 base pairs of terminal sequence identity and generates overhangs by using a proprietary 5’ exonuclease. The complementary 3’ overhangs are subsequently filled in with a DNA polymerase upon annealing. DNA ligase is used on the final product to seal nicks and create a continuous assembly. The entire reaction is incubated at a temperature of 50°C for as little as 15 minutes in a recommended volume of 20 μL.
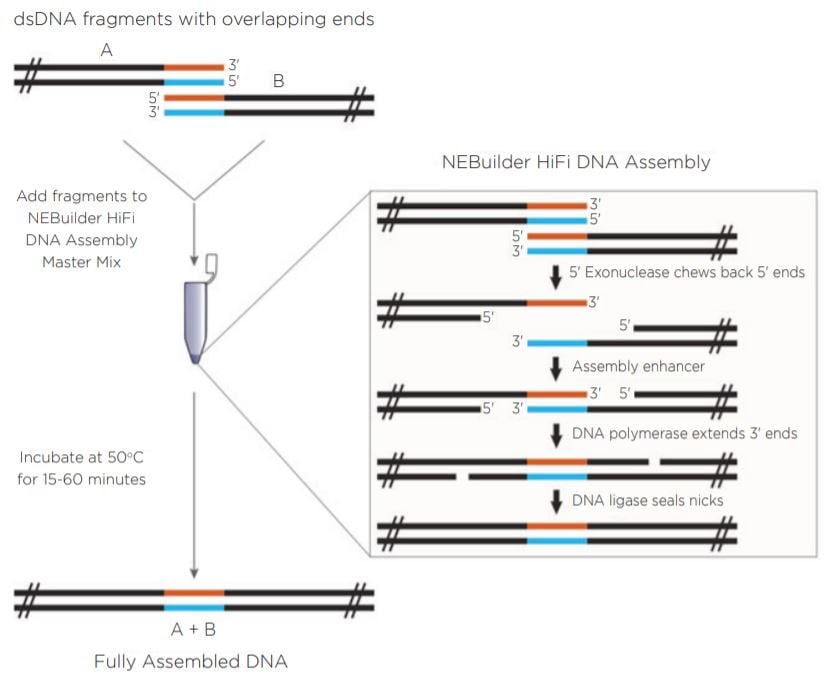 |
Previous work has shown the ability of the Echo 550 Liquid Handler to generate two-piece assemblies using the Golden Gate and Gibson Assembly chemistries utilizing a miniaturized protocol.1 The Echo 550 Liquid Hander uses a transducer to acoustically dispense in increments of 2.5 nL. In this experiment, NEBuilder HiFi chemistry was combined with the 25 nL increment dispensing Echo 525 Liquid Handler. Figure 1: Reaction progression of input DNA using the NEBuilder HiFi Master Mix. Reprinted from www.neb.com (2018) with permission from New England Biolabs, Inc. |
|
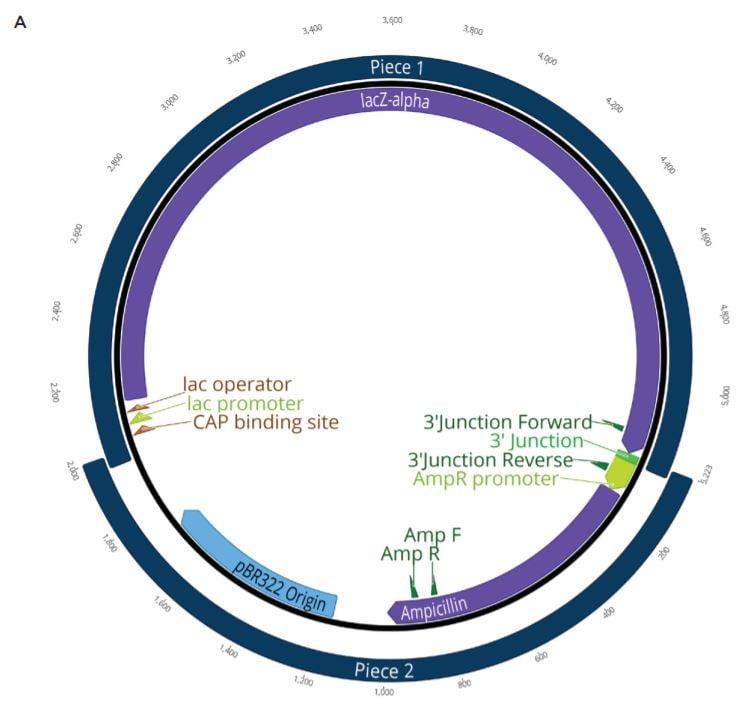
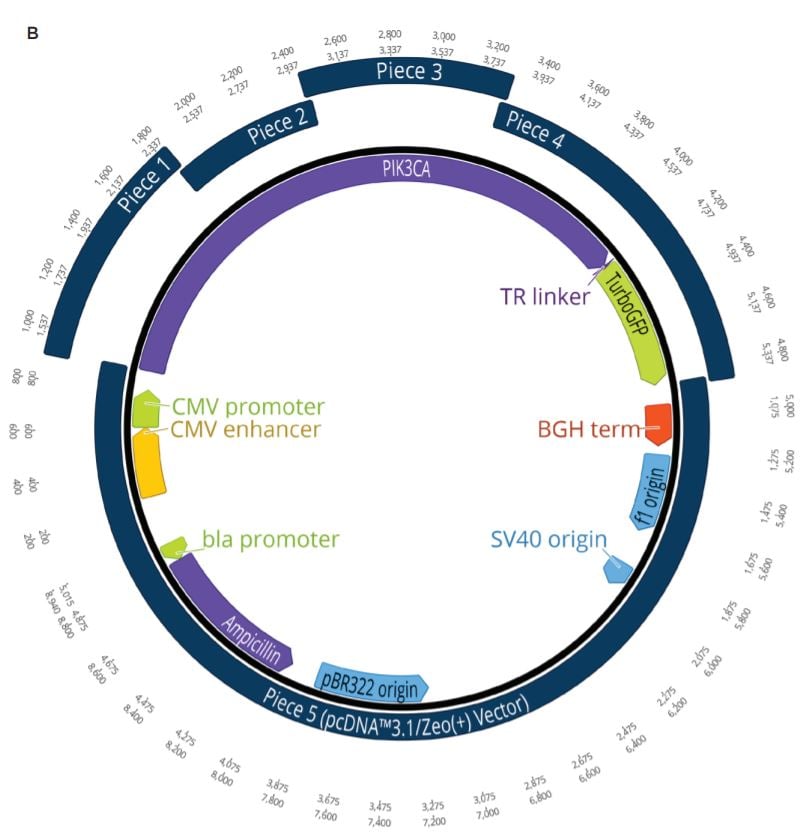
Two unique assemblies were designed. The first assembly contained two pieces as shown in Figure 2A. This construct was designed to reproduce the LacZ pathway in a 5223 bp plasmid. This would allow bluewhite screening for correctly assembled plasmids when transformed into a non-alpha complementing E. coli strain. A second, five-piece assembly with 8,940 bp was designed to more closely represent the complexity and size of typical industrial DNA construction (Figure 2B). The goals of these experiments were to utilize the Echo 525 Liquid Handler in assembling both a two-piece and five-piece assembly in reduced volumes. The Echo Liquid Handler enables lower-cost methods and workflows to produce high-quality synthetic DNA which expands design-based testing with higher throughput and affords the scientist a broader biological landscape to interrogate. Ultimately, the utilization of the Echo Liquid Handler for synthetic biology can help close the DNA read-write cost gap. Figure 2: A: (top) Plasmid Map for the two-piece assembly construct showing the plasmid replicative machinery and lacZ operon on the two pieces. The designed overlaps can also be seen where the pieces occupy the same plasmid locations. The area labeled 3’ junction is the area tested for by follow up qPCR whose primers are also shown in green. B: (bottom) Plasmid Map for the five-piece assembly construct showing the plasmid replicative machinery and split PIK3CA gene in relation to the five pieces. The designed overlaps can also be seen where the pieces occupy the same plasmid locations. These overlaps have also been called junctions and are labeled as ligation points in light green. |
Two-Piece Assembly
Rational Construct Design
The two-piece positive control included in the NEBuilder HiFi kit was used to miniaturize the reaction volume for DNA assembly. The NEBuilder HiFi kit positive control consists of four DNA fragments from two digested plasmids. The first is pUC19 that has been cut with AatII and HindIII to drop out the lacZalpha fragment. The second plasmid is CamR, digested to drop out a full length lacZ gene. The full length lacZ gene has been engineered to contain overlaps with pUC19 cut with AatII and HindIII. Upon addition of the NEBuilder HiFi Master Mix, the overlapping pieces will be chewed back, anneal, and ligate creating a new plasmid containing the replicative machinery from pUC19 along with the full lacZ operon from CamR. This plasmid will be also Ampicillin resistant. When transformed into a non-alpha complementing E. coli strain, such as NEB T7 Express, only constructs with the correct assembly will turn blue when induced with IPTG and the X-gal carbon source. This allows for an effective blue white screen to test for correct assemblies.
Echo Liquid Handler Assembly
Both the two-piece DNA mixture and the NEBuilder HiFi Master Mix were separately added into an Echo Qualified 384-Well Low Dead Volume Plus Microplate (384 LDV Plus). The NEBuilder HiFi protocol has a recommended total assembly volume of 20 μL with 10 μL of the two-piece assembly and 10 μL of the NEBuilder HiFi Master Mix. The volumes dispensed for each reaction size are shown in Table 1. The NEBuilder HiFi Master Mix was arrayed into a 384-well Bio-Rad PCR plate in a seven-fold (1500 nL) to four hundred-fold (25 nL) reduced volume. The NEB Positive control was then added in an equal volume. After arraying and sealing, the reaction was conducted for 30 minutes at 50°C in an Applied Biosystems ProFlex PCR System with a heated lid. Each reaction was done in triplicate in two separate PCR plates to allow for downstream junction qPCR and transformation.
NEBuilder HiFi Two-Piece Reaction Volumes
| 3000 nL Reaction (nL) | 2000 nL Reaction (nL) | 1000 nL Reaction (nL) | 500 nL Reaction (nL) | 250 nL Reaction (nL) | 150 nL Reaction (nL) | 100 nL Reaction (nL) | 50 nL Reaction (nL) | Echo Calibration | |
| NEB Positive Control Two-Piece Assembly | 1500 | 1000 | 500 | 250 | 125 | 75 | 50 | 25 | 384LDV_ PIus_AQ_ GP |
| NEBuilder HiFi Master Mix | 1500 | 1000 | 500 | 250 | 125 | 75 | 50 | 25 |
30 minutes 50° C incubation
Table 1: Volumes of reagents used for NEB two-piece miniaturization. The total reaction volumes ranged from 3000nL to 50nL. The twopiece assembly DNA and the NEBuilder HiFi Master Mix were transferred with the Echo 525 Liquid Handler from a 384 LDV Plus plate. These reactions were used in both the junction qPCR (Tables 2-3, Figure 1) and for E. coli transformation (Table 4).
Junction qPCR Testing
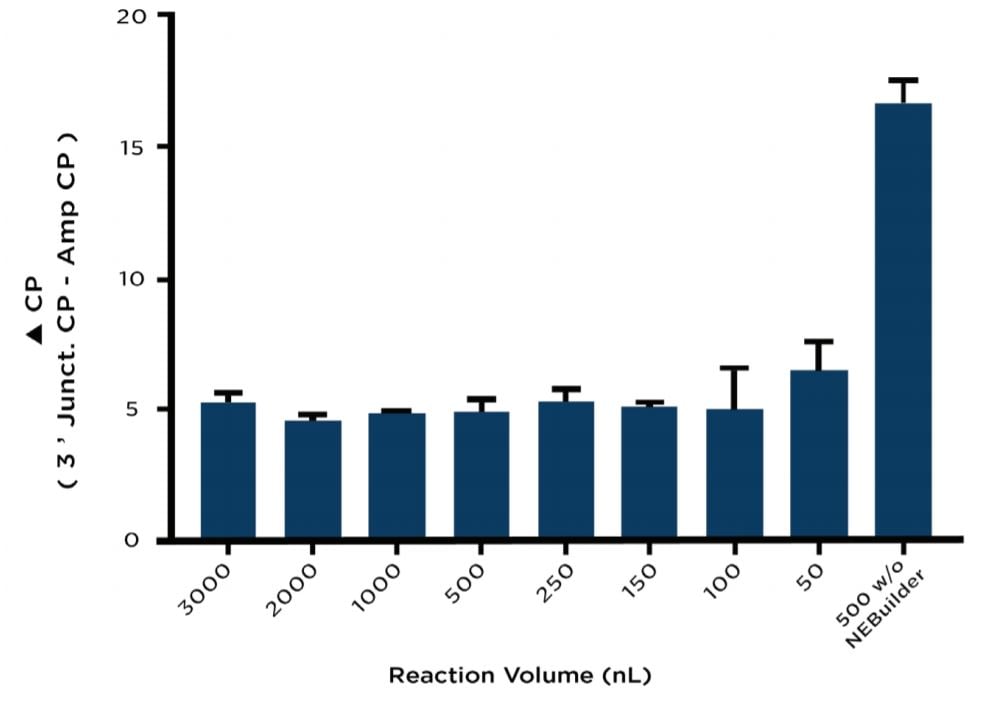
As previously mentioned, duplicate assembly plates containing different reaction volumes were tested in triplicate with one plate being reserved for assembly verification by junction qPCR upon reaction completion. The reactions on this plate were all normalized to the maximum reaction volume of 3000 nL using diH2O dispensed from an Echo Qualified 384-Well Polypropylene Microplate. The normalized NEBuilder HiFi reactions were then split into two wells each. For one of the split samples, a 100 bp region from the Ampicillin marker was amplified as a control gene to normalize the variable input DNA for each condition. A 161 bp fragment from the 3’ junction shown in Figure 2A was amplified from the complementary well of the split sample to test the NEBuilder HiFi DNA assembly. At the 500 nL volume, a control against overlapping PCR was included by using water instead of the NEBuilder HiFi Master Mix. Reagents were added according to Table 2 and run on a Roche LightCycler 480 according to the conditions outlined in Table 3. When normalized for DNA concentration by subtraction of the Ampicillin control amplicon, the reactions between 3000nL and 150 nL with NEBuilder HiFi occurred with a ΔCP ranging between 4.5 and 5.5 as compared to a ΔCP of 16.5 for the NEBuilder HiFi absent control (Figure 3).
Junction qPCR Reaction Volumes
|
Table 2: Reagent additions used for junction qPCR reactions. The Forward and Reverse Primers were designed to amplify either the 3’ junction or subset of the Ampicillin marker as seen in Figure 2A. |
Colony qPCR Reaction Conditions
|
Table 3: Roche Lightcycler 480 conditions run for the two-piece junction qPCR targeting the 3’ junction and Ampicillin control. |
Junction qPCR
Transformation / Construct Production
The duplicate plate of the two-piece NEBuilder HiFi assembly was transformed into 10 μL chemically competent NEB T7 E. coli following the conditions outlined in Table 4. The transformants were manually transferred into a 384-Well LDV Plus plate from their 384-well PCR plate after cooling and gently mixed by pipetting. In addition to the eight reaction volumes, a pUC19 control plasmid was also transformed. LB Agar OmniTrays with 100 µg/mL carbenicillin, 60µg/ml X-Gal, and 0.1mM IPTG were warmed and dried in the 37°C incubator for one hour. The Echo calibration 384LDV_Plus_AQ_GP was used to array 9 columns of 25 nL per construct transformation onto two separate OmniTrays. The plates were inverted and incubated overnight at 37°C.
NEBuilder HiFi Two-Piece Reaction Transformations
| 3000 nL Reaction (nL) | 2000 nL Reaction (nL) | 1000 nL Reaction (nL) | 500 nL Reaction (nL) | 250 nL Reaction (nL) | 150 nL Reaction (nL) | 100 nL Reaction (nL) | 50 nL Reaction (nL) | |
| NEB T7 E. coli | 10000 | 10000 | 10000 | 10000 | 10000 | 10000 | 10000 | 10000 |
30 minutes 50° C incubation
30 seconds 42°C incubation
2 minutes 4°C incubation
Table 4: Transformation conditions for the two-piece assemblies. A volume of 10 μL of NEB T7 E. coli was used for all reaction volumes. After the second 4°C incubation, the cells were transferred into a 384-well Echo LDV Plus plate and plated onto LB+Carb OmniTrays using acoustic liquid dispensing. No recovery time or media was included, as opposed to NEB heat-shock protocol.
Sanger Sequencing Verification
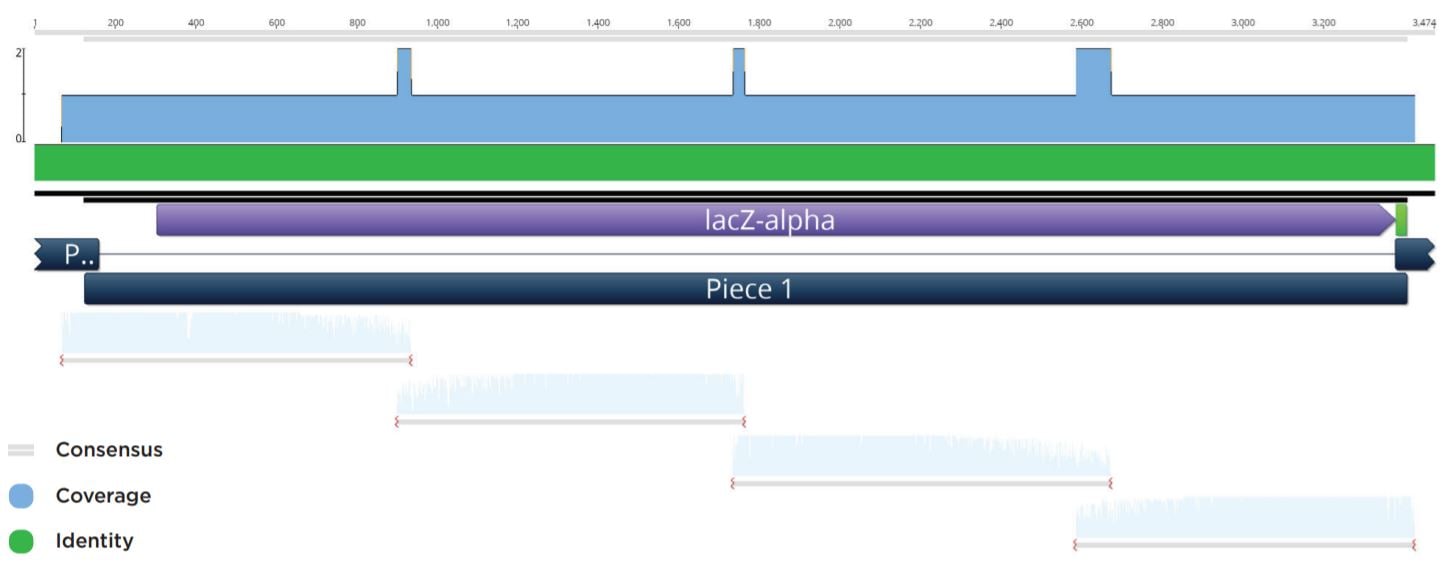
Blue colonies were observed at volumes down to 100 nL for the previous reactions. Representative clones were subsequently picked and used to start 4 mL cultures in LB Carbenicillin broth in 15 mL cell culture tubes. The biomass from these cultures were pelleted before being processed through a Qiagen Mini-prep kit and eluted in 50 μL Elution Buffer. The plasmid preparations were then submitted to Quintara Biosciences for Sanger sequencing. The Sanger sequencing was aligned to the reference assembly by Geneious as seen in Figure 4.
Figure 4: Representative sequencing alignment demonstrating the correct full sequence coverage of the LacZ operon within the twopiece assembly with overlapping coverage of vector.
Five-Piece Assembly
Rational Construct Design
A five-piece assembly was also designed to be used in the miniaturized reaction volumes to more closely simulate the challenges faced in real-world applications. The PIK3CA cancer gene was chosen to be broken up into four component pieces and constructed into the pcDNA3.1/Zeo(+) mammalian vector. This vector is interchangeably referred to as piece 5 after being linearized with KpnI-HF. All of the insert pieces were designed with 23 to 25 base pair DNA overlaps and ordered from Integrated DNA Technologies as gBlocks Gene Fragments. The vector and gBlocks were amplified to obtain the necessary amount of modular DNA required for the study.
Modular DNA Component Production (gBlocks Pieces 1-4)
The primers listed in Table 5 were ordered from Integrated DNA Technologies (IDT) to generate additional copies of the modular DNA for assembly. The high-fidelity polymerase Q5 was used for the PCR reactions as shown in Table 6. Each reaction was set up in a 1.5 mL tube, gently mixed, and aliquoted into a 384-well Bio-Rad PCR plate. The reactions were then run on an Applied Biosystems ProFlex PCR System with the reaction conditions shown in Table 7.
gBLOCK Amplification Primers
| Piece # | Forward Primer Sequence | Reverse Primer Sequence |
| Piece 1 | TAGCGTTTAAACTTAAGCTTGGTACATGCCTC | AAGATGGCATTGTAAAACAGTCCATTGGC |
| Piece 2 | AGAAAGCCTTTATTCTCAACTGCCAATGGAC | CTGCGTGGGAATAGCTAAATCCTGCTTC |
| Piece 3 | GCAGGATTTAGCTATTCCCACGCAG | GACATGATGTCTGGGTTCTCCCAATTCAA |
| Piece 4 | GGACTAGTGGATCCGAGCTCGGTAC | ATTGGGAGAACCCAGACATCATGTCAGAG |
Table 5: Primer sequences used to PCR amplify the gBlock template to increase concentration of necessary DNA for subsequent DNA assembly.
Piece PCR Reaction Conditions
| Reagent | Volume (µL) Per Reaction |
| Forward Primer (10 μM) | 10 |
| Reverse Primer (10 μM) | 10 |
| NEB Q5 Polymerase 2x MM | 100 |
| IDT gBlock DNA (1ng/μl) | 2 |
| diH2O | 78 |
Table 6: Volumes of reagents per PCR reaction used for template DNA expansion. The total reaction was dispensed in 20 μL aliquots into a 384-well Bio-Rad PCR plate. Q5 polymerase was chosen due to the need for a low error rate with the amplified pieces for downstream sequence fidelity.
Thermocycler Reaction Conditions
| Piece # | Initial Denaturization | 25 Cycles | Final Extension | Hold | ||
| Time | 30 minutes | 10 minutes | 30 minutes | 1 hour | 2 hours | infinite |
| Temperature | 98°C | 98°C | 71°C | 72°C | 72°C | 4°C |
Table 7: Proflex thermocycler reaction conditions used for the amplification of the insert piece DNA (Pieces 1-4). The reactions were held to 25 cycles to minimize the number of mutations generated.
Vector DNA Component Production (pcDNA3.1/Zeo (+) Piece 5)
The circularized mammalian vector pcDNA3.1/Zeo (+) was purchased from ThermoFisher Scientific and transformed into NEB 10-beta competent E. coli. A clonal transformant was frozen in glycerol at -80°C for use in the entirety of the experiment. An overnight 25 mL culture was set up in a 250 mL Erlenmeyer Flask in LB Broth with 100 µg/mL carbenicillin and incubated at 37°C. It was split between 5 Qiaprep columns using the manufacturer recommended protocol and eluted in 250 μL. The eluted plasmid DNA was then digested overnight at 37°C with the addition of 20 μL NEB KpnI-HF and 30 μL 10x Fast Buffer.
DNA Cleanup, Quantitation, and Visualization (Pieces 1-5)
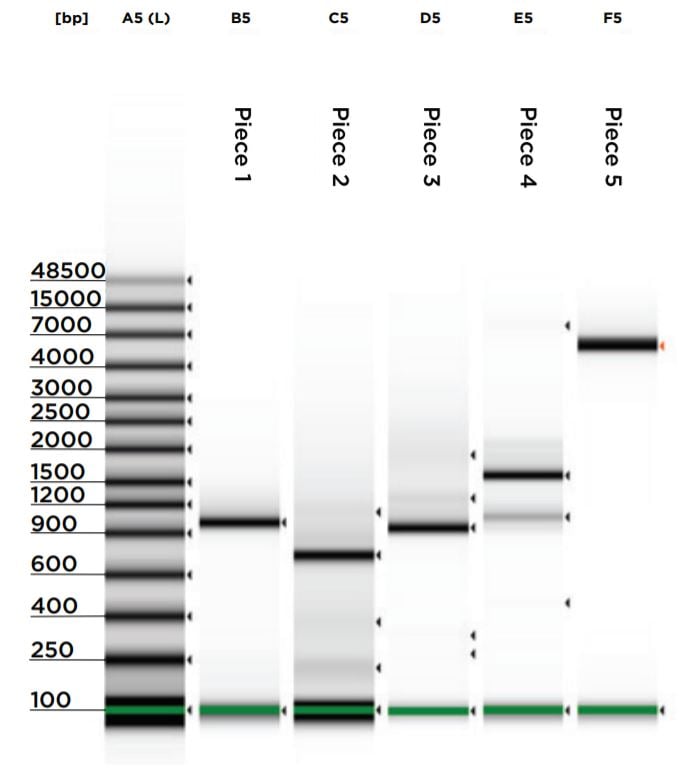
Upon reaction completion, both the vector restriction digest and gBlock PCRs were cleaned and concentrated using the NEB Monarch PCR & DNA Cleanup Kit (5 μg). The kit was processed according to the kit protocol and eluted in 11 μL of elution buffer. A 1 μL aliquot for each reaction was run on an Agilent 2200 Tapestation loaded with a Genomic DNA Analysis ScreenTape using their given protocol (Figure 5). The DNA was visually checked for quality, and the concentration of the expected band size was used as input for DNA normalization in DNA assembly.
DNA and NEBuilder HiFi Master Mix Arraying
Using the DNA concentrations from the Agilent 2200 Tapestation run in Figure 5, the DNA was diluted to be within a desirable range for assembly (Table 8). It was then transferred into an Echo Qualified 384 LDV Plus plate. NEBuilder HiFi Master Mix was also separately added to the 384 LDV Plus plate to allow for a low dead volume of enzyme mix. The NEBuilder HiFi protocol has a recommended assembly volume of 20 μL with an input of 0.5 pmol per piece for complex assemblies. The NEBuilder HiFi Master Mix was arrayed into a 384-well Bio-Rad PCR plate in a five-fold (4000 nL) to eighty-fold (250 nL) reduced volume. The DNA was then added to each reaction maintaining the molecular end ratio for each piece. Note that the minimal transfer volumes of DNA were attained in the 250nL reaction. After arraying and sealing, the reaction was conducted for 1 hour at 50°C in an Applied Biosystems ProFlex PCR System with a heated lid. The PCR plate was cooled to 4°C before being transformed into NEB 10B E. coli cells.
NEBuilder HiFi Five-Piece Assembly Reaction Volumes
| Diluted DNA Concentratration (ng/µL) | 4000 nL Reaction (nL) | 3000 nL Reaction (nL) | 2000 nL Reaction (nL) | 1000 nL Reaction (nL) | 750 nL Reaction (nL) | 500 nL Reaction (nL) | 250 nL Reaction (nL) | Echo Calibration | |
| Piece 1 | 20 | 325 | 250 | 150 | 75 | 50 | 50 | 25 |
384LDV_PIus_AQ_GP |
| Piece 2 | 20 | 225 | 175 | 100 | 50 | 50 | 25 | 25 | |
| Piece 3 | 20 | 275 | 200 | 150 | 75 | 50 | 25 | 25 | |
| Piece 4 | 50 | 200 | 150 | 100 | 50 | 50 | 25 | 25 | |
| Piece 5 | 100 | 325 | 250 | 175 | 75 | 50 | 50 | 25 | |
| diH2O | 650 | 475 | 325 | 175 | 125 | 75 | 0 | ||
| NEBuilder HiFi Master Mix | 2000 | 1500 | 1000 | 500 | 375 | 250 | 125 |
1 hour 50°C incubation
Table 8: Volumes of reagents used for NEB 5-piece miniaturization. The total reaction volumes were transferred by the Echo 525 Liquid Handler down to 250 nL. Due to the increased number of pieces assembled in comparison to the two-piece assembly, 250 nL was the minimum volume that could be attempted. These reagents were all dispensed from a 384 LDV Plus plate. These reactions were all transformed into E. coli.
Transformation / Construct Production
The five-piece reactions for the NEBuilder HiFi assembly miniaturization were transformed into chemically competent NEB 10B E. coli following the conditions outlined in Table 9. The volume of cells listed in the table was pipetted into the 384 PCR plate containing the reactions that had been chilled to 4°C. After a 30-minute incubation for the mixture at 4°C, the cells were heat shocked at 42°C for 30 seconds then immediately transferred back to the 4°C cold block. The transformants were then manually transferred into a 384-Well LDV Plus plate from the 384-well PCR. LB Agar OmniTrays with 100 µg/mL carbenicillin were warmed and dried in the 37°C incubator for 1 hour. The Echo calibration 384LDV_Plus_AQ_GP was used to array 8 columns of 25 nL per construct transformation onto OmniTrays. The plates were inverted and incubated overnight at 37°C. Isolated colonies were subsequently used for verification by colony qPCR and Sanger sequencing.
NEBuilder HiFi Five-Piece Reaction Transformations
| 4000 nL Reaction (nL) | 3000 nL Reaction (nL) | 2000 nL Reaction (nL) | 1000 nL Reaction (nL) | 750 nL Reaction (nL) | 500 nL Reaction (nL) | 250 nL Reaction (nL) | |
| NEB 10B E. coli | 40000 | 40000 | 40000 | 25000 | 18750 | 12500 | 6250 |
30 minutes 4°C incubation
30 seconds 42°C incubation
2 minutes 4°C incubation
Table 9: Transformation conditions for the five-piece assembly miniaturization reactions. A variable volume of 40 µL to 6250 nL NEB 10B E. coli was used. 40 µL was the maximum that could be contained in the 384-well PCR plates used for transformation. After the 4°C cooling step, the cells were transferred into a 384-well Echo LDV Plus plate and plated onto LB+Carb OmniTrays using acoustic liquid dispensing.
Colony qPCR Quality Control
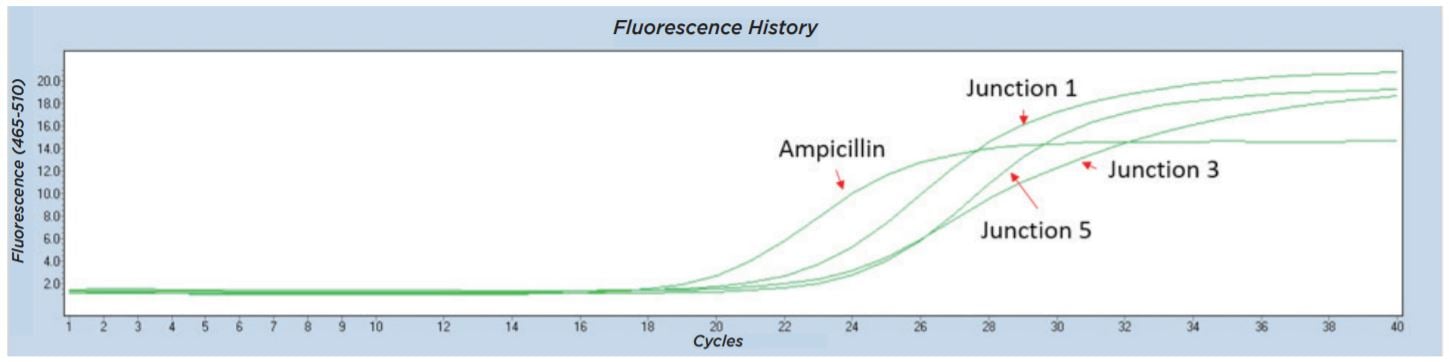
The Echo 525 Liquid Handler was used to accurately dispense 250 nL of a cell suspension into four 10 μL qPCR reactions targeting four different junction regions of the assembled plasmid construct (Figure 6). A qPCR reaction master mix was made for each of the four reactions containing the amplicon primer pair and LightCycler 480 SYBR Green I Master as shown in Table 10. The reagent mixture minus the colony suspension was bulk dispensed into a barcoded Bio-Rad Hard-Shell Skirted 384-Well PCR Plate. After reagent addition, the qPCR plate was centrifuged for 1 minute at 1000 × g. Isolated colonies were picked using a P10 pipette tip into 50 μL of diH2O in an Echo Qualified 384-Well Polypropylene Microplate 2.0 (384-Well PP). The cell-containing PP plate was sealed and briefly mixed at 2000 RPM for 2 minutes on an Eppendorf MixMate. From this, 250 nL of the cell suspension was dispensed using the Echo calibration 384PP_AQ_BP into each of the four qPCR reagent mixes. The qPCR plate was then sealed with optical film, vortexed for 2 minutes at 2000 RPM on the Eppendorf MixMate, and briefly centrifuged at 1000 × g for 1 minute. The colony qPCR was run according to the conditions in Table 11 on a Roche Lightcycler 480. Colonies with all three junction CPs closely matched to the Ampicillin control were called positive assemblies and moved to Sanger sequencing. An example trace of an assembly that was called as a positive is shown in Figure 7.
Colony qPCR Reaction Volumes
| Reagent | Volume (nL) per reaction | Echo Calibration |
| Forward Primer (10 µM) | 250 | |
| Reverse Primer (10 µM) | 250 | |
| LightCycler 480 SYBR Green I Master | 5000 | |
| diH2O | 4250 | |
| Colony Cell Suspension | 250 | 384PP_AQ_BP |
Table 10: Reagent additions used for the five-piece colony qPCR reactions. Four different primer pairs were used to amplify the target regions from the colonies. Note that the primers, SYBR Green Master Mix, and water were made as a master mix and dispensed together using a pipette. 250 nL of the cell suspension was then dispensed into each of the four different reaction conditions using the Echo 525 Liquid Handler.
Colony qPCR Reaction Conditions
| Temp. (°C) | Hold (sec) | Ramp Rate °C/sec | Acquisition (465 nm/520 nm) | Cycles | |
| Pre-incubation | 95 | 300 | 4.8 | 1 | |
| Amplification | 95 | 10 | 2.5 | 40 | |
| 50 | 10 | 2.5 | Single | ||
| 72 | 10 | 2.5 | |||
| Melting Curve | 95 | 5 | 4.8 | 1 | |
| 65 | 60 | 2.5 | |||
| 97 | continuous | 0.11 | |||
| Cooling | 4 | 30 | 2.5 | 1 |
Table 11: Roche Lightcycler 480 conditions run for the five-piece junction qPCR looking for three generated junctions in comparison to the Ampicillin control.
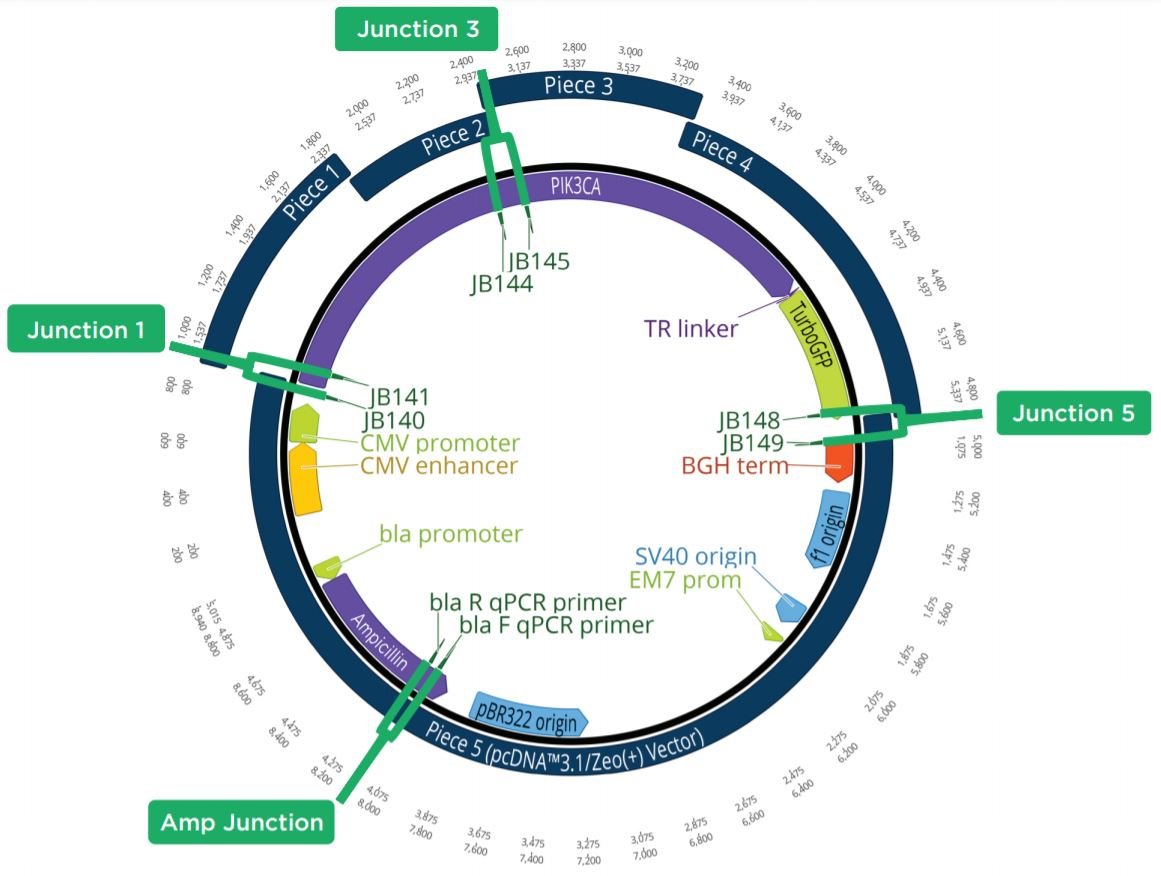
Figure 6: Plasmid Map showing the five-piece assembly construct with the 4 targeted qPCR amplicons highlighted. If Junctions 1, 3, and 5 had a similar CP to the Ampicillin control, the colony was considered positive by colony qPCR. By testing these junctions, the presence of all five pieces could be seen.
Figure 7: Representative Lightcycler 480 trace from colony qPCR testing for junctions from five-piece assembly. All four amplicons including the three tested junctions were found in this colony, so the sample was called a positive and moved forward for Sanger sequencing.
Sanger Construct Validation
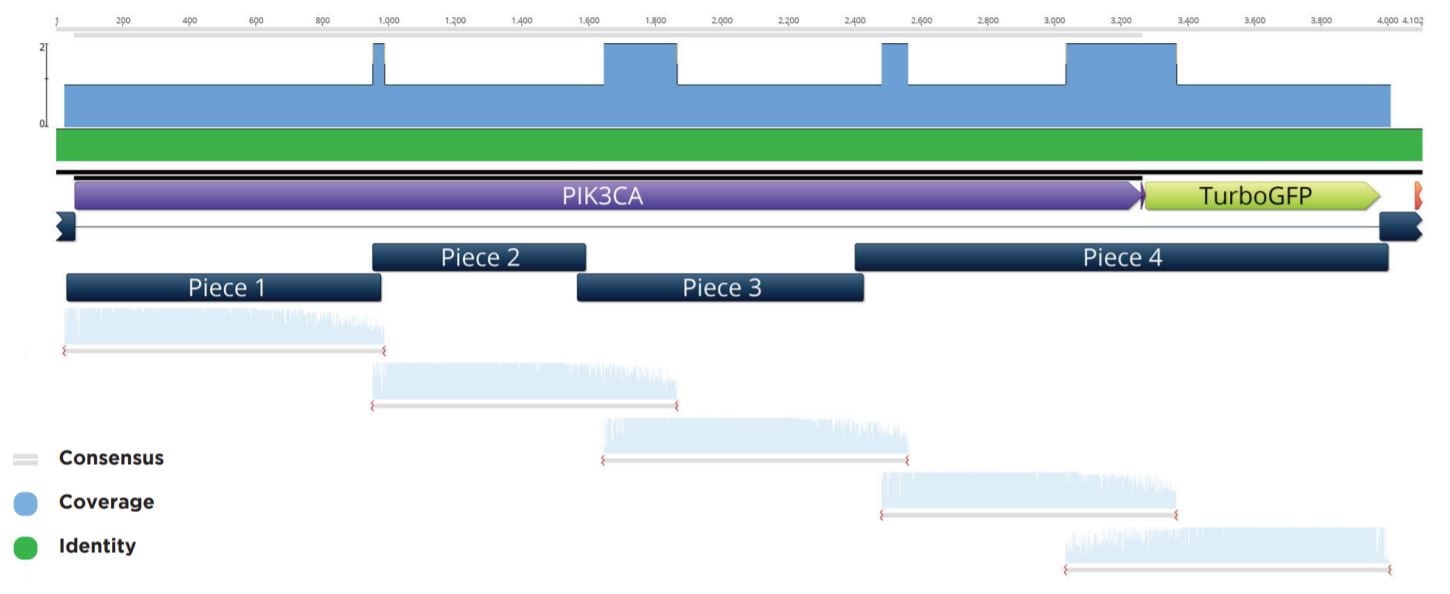
Colonies that passed the colony qPCR screening were spotted with 50 nL onto a fresh LB+Carb100 OmniTray from the 384-well PP plate used in the above qPCR QC step and grown overnight at 37 °C. These spots were used to start 6 mL cultures in LB Carbenicillin liquid media in 15 mL cell culture tubes. The biomass from these cultures was pelleted before being processed through a Qiagen Miniprep kit and eluted in 50 μL Elution Buffer. The plasmid preparations were then submitted to Quintara Biosciences for Sanger sequencing. The Sanger sequencing was aligned to the reference five-piece assembly by Geneious as seen in a representative alignment in Figure 8.
Figure 8: Representative sequencing alignment demonstrating the correct full sequence coverage of the PIK3CA gene within the five-piece assembly. Note that five overlapping Sanger sequencing reactions were needed to obtain full coverage for the inserted modular pieces.
Discussion
Reducing overall reagent costs while maintaining the robustness of the DNA construction pipeline is a goal of many research facilities to allow for a greater number of testable hypotheses. By combining the NEBuilder HiFi kit with the Echo 525 Liquid Handler this study has achieved those aims over the standard DNA construction workflow in numerous ways. The miniaturization and workflow enhancements were initially demonstrated in a less challenging two-piece assembly before being moved into a more challenging five-piece modular assembly.
The first project goal reached was the successful 200-fold reduction in volume of a two-piece LacZ operon assembly. This assembly was accomplished at a 100 nL total volume from a 20 μL protocol recommended volume. The LacZ operon was successfully expressed in these constructs for a telling blue coloration when plated on the X-Gal carbon source with IPTG induction in NEB T7 E. coli cells. The acoustic plating of these cells onto OmniTrays was also a workflow enhancement with the potential to automate a time-consuming and space intensive step in many protocols. The constructs were also verified by Sanger sequencing with the blue clones containing the expected sequence as shown in Figure 3. As shown in this study with the NEBuilder HiFi kit and previous other chemistries, DNA construction is robust for two-piece assemblies in the nanoliter range.
The other main project goal was the reduction in volume of a complex modular assembly more representative of a typical industrial DNA construct. For this modular assembly, positive clones were observed with a reduction of eighty-fold with the most robust results occurring at the 500 nL or fortyfold reduction volume. At the 500 nL reduced volume, only 1.25 fmoles of DNA was used for each input piece, which would dramatically decrease the amount of DNA required from that of the recommended protocol. The input DNA is an often overlooked cost and can be highly variable between experiments due to synthesis cost and in-house preparation. The five-piece assembly utilized modular DNA pieces, which makes the conservation of input DNA crucial as they are reused over many different assemblies. As mentioned with the two-piece assembly, OmniTray plating was also a noteworthy time-saver and decreased overall incubator space requirements. Finally, colony junction qPCR allows for a rapid analysis of the success of a reaction as well as the removal of background constructs. The colony qPCR quality control step was a method developed to screen incorrect colonies prior to being further moved down the sequence validation pipeline. No background vector carryover was detected in those samples that passed the colony qPCR step, which is a commonly encountered problem. A final improvement from standard workflows is the ability to work in a 384-well footprint throughout the experiment as opposed to a 96-well format, which represents an obvious four-fold advantage in throughput. The ability of the Echo Liquid Handler to seamlessly integrate with standard SBS footprint plates due to tipless liquid handling allows both the use of OmniTrays for plating and the 384-well PCR plates used for the NEBuilder HiFi reactions. The aim of miniaturization for a five-piece modular assembly was reached by the Echo 525 Liquid Handler with a robust forty-fold reduction in volume along with significant secondary workflow enhancements.
Working at the nanoliter scale in synthetic biology would have been inconceivable in the recent past, but the Echo Acoustic Technology makes these volumes a reality. Plasmid constructs created using the NEBuilder HiFi kit were able to be reduced 200-fold in volume, which will significantly decrease the cost of DNA assembly allowing for substantial increases in throughput. The integration of a Echo 525 Liquid Handler into a DNA construction pipeline, such as one using the NEB NEBuilder HiFi kit, allows for significant volume reductions as well as workflow enhancements with both complex and simple plasmid designs.
Reference
1. Kanigowska P, Shen Y, Zheng Y, Rosser S, Cai Y. Smart DNA Fabrication Using Sound Waves: Applying Acoustic Dispensing Technologies to Synthetic Biology. J Lab Autom. 2015:1-8. doi:10.1177/2211068215593754.
Materials
| Equipment | Manufacturer |
| Echo 525 Liquid Handler | Beckman Coulter Life Sciences |
| Allegra X-14 Centrifuge | Beckman Coulter Life Sciences |
| MixMate | Eppendorf |
| TapeStation 2200 | Agilent |
| Incu-Shaker Mini, Shaking Incubator | Benchmark Scientific |
| ProFlex PCR System | Thermo Fisher |
| LightCycler 480 | Roche |
| Reagents | Manufacturer | Part Number |
| DNA Assembly Cloning Kit | New England BioLabs | #E5520 |
| KpnI-HF | New England BioLabs | #R3142 |
| Q5 High-Fidelity 2X Master Mix | New England BioLabs | #M0492 |
| QIAprep Spin Miniprep Kit | QIAGEN | #27104 |
| Genomic DNA Analysis ScreenTape Kit | Agilent | #5067-5365, #5067-5366 |
| NEB 10-beta Competent E. coli (High Efficiency) | New England BioLabs | #C2019l |
| T7 Express Competent E. coli (High Efficiency) | New England BioLabs | #C2566l |
| LB Broth with 100 µg/mL Carbenicillin | Teknova | #L8185 |
| LB Agar OmniTrays with 100 µg/mL Carbenicillin | Teknova | #L2010 |
| LB Agar OmniTrays with 100 µg/mL Carbenicillin, 60µg/ml X-Gal, and 0.1mM IPTG | Teknova | #L2906 |
| gBlocks Gene Fragments | Integrated DNA Technologies | Custom DNA |
| Junction qPCR Primers | Integrated DNA Technologies | Custom Oligos |
| Piece Amplification Primers | Integrated DNA Technologies | Custom Oligos |
| LightCycler 480 SYBR Green I Master | Roche | #04707516001 |
| pcDNA 3.1/Zeo (+) Mammalian Expression Vector | Thermo Fisher Scientific | #V86020 |
| Monarch PCR & DNA Cleanup Kit (5 μg) | New England BioLabs | #T1030S |
| 200 Proof Ethanol | Sigma Aldrich | #E7023 |
| Consumables | Manufacturer | Part Number |
| Echo Qualified 384-well PP Microplate | Beckman Coulter Life Sciences | 001-14555 |
| Echo Qualified 384-well LDV Plus Microplate | Beckman Coulter Life Sciences | 001-12782 |
| TapeStation Plate | Agilent | #5067-5150 |
| 14 mL Round Bottom Snap Cap Polypropylene Tubes | EK Scientific | EK-62261 |
| Hard-Shell Skirted 384-Well PCR Plate | Bio-Rad | #HSP3805 |
| Hard-Shell 384-Well PCR Plates, Barcoded | Bio-Rad | #HSP3901 |
| Microseal 'C' PCR Plate Sealing Film | Bio-Rad | #MSC1001 |
| 384 well Plates, Polystyrene Black (with Micro-Clear Bottom) | Greiner | #781096 |
| Nalgene Single-Use PETG Erlenmeyer Flask (250 mL) | Thermo Fisher | #4112-0250 |
| 1.5 mL DNA LoBind Tubes | Eppendorf | #022431048 |