A General Guide to Lipid Nanoparticles
Introduction
Lipid-based drug delivery (LBDD) systems are highly versatile, and have been used to deliver various bioactive molecules to targeted cells and tissues. LBDD systems have several advantages over conventional drug delivery methods, including increased drug stability, higher bioavailability, and optimized drug distribution. Lipid nanoparticles (LNPs) are one type of LBDD system, and represent a significant advancement for the delivery of oligonucleotidebased vaccines and gene therapies. mRNA, siRNA, and pDNA are common oligonucleotide cargoes, but must withstand degradation by ribonucleases, renal clearance, and immune responses in order to function as therapeutics.1 Encapsulation inside LNPs can overcome these limitations, enabling the formulation to reach its target tissue and remain effective.2 mRNAcontaining LNPs now form the basis of most COVID-19 vaccines, protecting the payload from degradation within the body. LNPs are also used to deliver gene therapies such as patisiran, a transthyretin-targeting siRNA for the treatment of hereditary amyloidogenic transthyretin amyloidosis.1 In this case, the LNPs not only help to protect the therapeutic during circulation, but also enhance intracellular delivery into hepatocytes.
LNP composition
LNPs are typically composed of a monolayer shell made of ionizable cationic lipids, glycerophospholipids, sterol lipids, and PEGylated lipids, surrounding an internal core containing an aqueous phase and reverse micelles. Each structural component of an LNP plays a specific role in oligonucleotide-based drug encapsulation and delivery. Changing particle composition — for instance, by altering surface charge, size or lipid content — affects their delivery success and therapeutic efficacy.
Oligonucleotides are negatively charged, whereas ionizable cationic lipids possess a positive charge at low pH. This enables them to self-assemble into reverse micelles capable of encapsulating oligonucleotide cargoes. Ionizable cationic lipids have a near-neutral charge at the body pH, and so can deliver their oligonucleotide therapy without causing cytotoxicity.
Glycerophospholipids comprise a hydrophilic head group, which determines the overall LNP charge, and two hydrophobic fatty acyl tails attached to a glycerol backbone. Neutral phospholipids improve the efficacy of membrane fusion, and anionic lipids are typically used for the delivery of small molecules, and to prevent aggregation during storage.
Sterol lipids, such as cholesterol, are used to fill membrane packing defects, add structural integrity, aid in membrane fusion, and support the delivery of oligonucleotide cargoes.
PEGylated lipids prevent serum protein adsorption, which commonly inhibits drug delivery, and can also be used to improve cellular targeting and uptake. Incorporating PEGylated lipids and increasing particle size can improve LNP delivery to certain target organs.1
An overview of LNP manufacturing techniques
LNPs are most frequently prepared by combining an ethanolic lipid mixture with an acidic aqueous solution that contains oligonucleotides. There are a few ways to carry out ethanolic mixing for laboratory-scale LNP production, each with their advantages and disadvantages.
A

| Component | Function | |
|---|---|---|
 |
Cationic lipid |
Encapsulation Membrane fusion |
.jpg?rev=e15450de2c6646da9318da05f2ca8dc1) |
Cholesterol | Stabilization |
 |
Neutral lipid |
Stabilization Improve processing |
 |
PEG shielding lipid |
Stop aggregation Control PK |
B
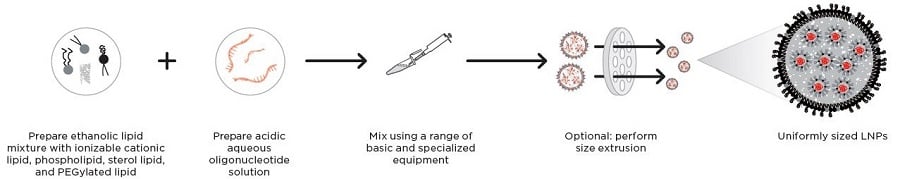
Figure 1: A: Scheme of LNP assembly including nucleic acid cargo and function of the four lipids B: Steps in LNP formation.
Automated microfluidic devices and microfluidic chips can be used to rapidly and reproducibly mix two streams of ethanolic lipid mixture and aqueous oligonucleotide solution to prepare homogeneous LNP formulations with high encapsulation efficiency. This option comes with higher capital costs, but provides greater scalability.
Ethanol injection into the aqueous oligonucleotide solution under continuous mixing — with a magnetic stirrer or by rapid hand pipetting — is a low cost option, but tends to yield more heterogeneous formulations than when using microfluidic devices and is less scalable.1
Ultrasonication, using sound waves with frequencies above 20 kHz, can also be used to homogenize LNP formulations during microfluidic manufacture.3 These high frequency pressure waves generate microbubbles in the LNP emulsion, which then collapse to cause cavitation — areas of localized high temperatures and pressure. This breaks down larger particles into smaller LNPs suitable for drug delivery. Ultrasonication can be used to tune nanoparticle size, enhance encapsulation efficiency, and improve drug release profiles, including bioavailability. It also prevents clogging of microfluidic chips and improves mixing efficiency and process stability.4
Characterizing LNPs
Parameters
Several parameters affect the efficacy of an LNP as a drug delivery system, and each must be characterized and adjusted to ensure optimum encapsulation and release of therapeutic cargo.2,5
- Lipid composition: The ionizable lipid nitrogen:oligonucleotide phosphate (N:P) molar ratio. This influences particle size, polydispersity, and therapeutic efficacy. Clinically relevant LNP formulations often contain an N:P of 3-6:1.1,5
- Encapsulation efficiency: The lipid:oligonucleotide weight ratio. This influences encapsulation efficiency — the proportion of total oligonucleotides taken into the LNPs. Most LNPs are formulated with a lipid:oligonucleotide weight ratio of 10:1.1 Microfluidic mixing yields formulations with the highest encapsulation efficiencies and the lowest percentage of empty LNPs.
- Copy number: The number of nucleic acid strands, for example mRNA, encapsulated inside the LNP, also known as the payload capacity.2 Nucleic acid payload and distribution influence the pharmacodynamics, pharmacokinetics, and delivery efficiency of an encapsulated therapeutic. The toxicity and immunogenicity of free mRNA in particular mandates rigorous characterization of the final product’s mRNA payload and distribution. These factors can be improved by enhancing encapsulation efficiency.
- Size: The average particle diameter, typically ~20-100 nm.1 This affects how much therapeutic cargo can be encapsulated. It also alters the pharmacokinetics of the administered particle, as LNPs of less than 100 nm can easily pass through fenestrated endothelium to penetrate target tissues, whereas larger particles may fail to penetrate and can elicit an immune response.1 Smaller particles typically also have longer circulation and half-lives than larger ones, as they evade elimination by the immune system. The particle size is dependent on the preparation method and whether extrusion is carried out.
- Polydispersity: The polydispersity index (PDI) is a measure of the LNP size distribution, with smaller PDI values indicating homogeneity and larger values indicating heterogeneity. The PDI value ranges from 0.0 — for completely homogeneous formulations — to 1.0 for a highly polydisperse sample.6 A PDI of <0.3 is considered to be acceptable for LBDD systems and indicates a homogeneous population.6 PDI influences the ability of an LNP to accumulate in the target tissue, with lower PDI values resulting in safer, more stable, and more consistent delivery systems. Quantifying polydispersity is critical for process development and comparability. The PDI value can be reduced by altering lipid composition, optimizing the mixing rate, changing the preparation method, or by carrying out an additional post-preparation extrusion step.
- Particle concentration: The concentration of loaded LNPs in a formulation. Particle concentration affects drug bioavailability, and quantification is necessary to ensure comparable results between experiments during research and development workflows. LNPs may be concentrated by ultracentrifugation or diluted to reach a target concentration.
- Charge: The overall charge of the LNPs. Positively charged particles may elicit a stronger immune response, whereas LNPs with a shell made of ionizable cationic lipids have a near-neutral charge at the body pH, and so are unlikely to cause cytotoxicity or immunogenicity. Altering surface charge can also direct LNPs to specific target tissues, for example, intravenously administered LNPs with a net negative charge can be targeted to the spleen.
- Stability: The stability of LNPs is important for their storage and function, and affects the performance of the final pharmaceutical products. Average particle size, PDI, and lipid and cargo integrity are vital components contributing to the long-term stability of LNPs.1,6
- Hydrodynamic radius: The size of the particle including the hydration layer and PEG chains that extend from the lipid core into the solvent. Overloaded particles have the widest hydrodynamic radius, while empty particles display the smallest. Clinically relevant LNPs demonstrate a hydrodynamic radius that sits in between these two extreme values, making measurement essential to ensure efficacy.
Analysis
There are multiple analytical methods that can be used to characterize LNPs, some of which are often used in conjunction to provide a more complete picture of LNP structure and function.
1. Analytical ultracentrifugation (AUC)
A

B

C

Figure 2: A: Schematic of AUC instrumentation B: Top view of sample/ reference cell assembly C: Detection modules provide a plot of sample concentration against radial distance along the sector length. These plots are collected over time.
AUC is a solution-based separation technique that can be used to rapidly differentiate between empty and clinically relevant LNPs in a sample. Density matching, multi-wavelength, and fluorescence detection AUC approaches can be used to provide high resolution composition information for LNP formulations and characterize LNP loading with nucleic acids.5 Delivery systems (lipids) and cargo (RNA) have vastly different densities, which influence their sedimentation behavior. This can be exploited by density matching experiments in the presence of different D2O or other water isotope concentrations. AUC also takes advantage of the spectral differences between a light scattering lipid nanostructure and a nucleic acid cargo — which has a unique chromophore at ~260 nm — to achieve optical separation.5
AUC allows for the determination of the hydrodynamic radius based on bulk observation — as opposed to the particle radius provided by cryogenic transmission electron microscopy (cryo- TEM) — and does not produce an intensity-weighted size distribution, unlike the dynamic light scattering (DLS) method. The AUC technique also provides sedimentation and diffusion coefficients, while still offering high resolution and sensitivity for heterogeneous samples. These values can then be used to derive anisotropy, density, molar mass, and particle size.5 Subsequent deconvolution of molar mass into the fractions of lipid and mRNA will enable the calculation of mRNA copy number per capsid.2 Another particular strength of the AUC method is its versatility; the technique can be used to study proteins and nucleic acids, drug delivery vehicles, antibodies, and other biologics. It can also be used to directly characterize nanoparticles under biologically relevant conditions, reflecting those in which a therapeutic may be administered to a patient to provide valuable insights into the physiological function of an encapsulated drug or vaccine.5 AUC methods are well suited to characterizing mRNA loading ratio, sample purity, and the presence of free mRNA, and excel in terms of resolution and information content when compared to traditional methods, such as DLS and cryo-TEM. These approaches therefore represent an important alternative for the routine analysis and validation of LNP formulations used for therapeutic applications.
The technique does, however, have some limitations. The density matching algorithm assumes that the density of the solvent inside the particle is the same as that of the bulk solvent, which may become an issue in formulations containing high concentrations of excipients.2 Furthermore, the algorithm is very sensitive to the absorbance baseline in heterogeneous samples; when baselines do not precisely align between sedimentation velocity experiments performed in different buffers, any larger differences in partial specific volumes can cause slight shifts in boundary fractions. This can be minimized by carefully fitting the original sedimentation velocity experiments, making sure that any buffer absorbance is properly accounted for in the time invariant noise component. It may also be avoided by lowering the number of divisions along the boundary.5
2. Cryogenic transmission electron microscopy (cryo-TEM)
Cryo-TEM can be used to study the structure of LNPs at near-atomic resolution. It involves freezing the samples in a thin layer of vitreous ice to preserve their near-native structures and prevent the formation of ice crystals that can damage the particles. The frozen grid containing the vitrified specimen is then transferred to the cryo-electron microscope, which uses a beam of electrons instead of light to image the specimen. The electron beam passes through the frozen sample, and the interaction of electrons with the particles produces a projection image.7 Numerous high resolution images are collected from different angles, and a three-dimensional reconstruction is generated using advanced computational methods, providing detailed structural information about the LNP and its molecular arrangement.8
Cryo-TEM provides several strengths for LNP characterization, for instance, it enables particles to be imaged in a near-native hydrated state, as rapid freezing in vitreous ice prevents the formation of ice crystals, preserving the near-natural structure of the sample, without the need for chemical fixatives.9 Together with the technique’s low dose imaging approach — which minimizes radiation damage to the specimen — this allows integration with other downstream techniques to provide a more comprehensive understanding of particle structures, such as X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and cryo-electron tomography.
However, cryo-TEM does possess some drawbacks for LNP imaging. First, it only allows for the determination of the particle radius, rather than the hydrodynamic radius, which includes the PEG hydration layer associated with the LNPs.5 In addition, it can be difficult for cryo- TEM to differentiate empty LNPs from those containing the clinically relevant loading in some formulations due to the insufficient electron density of the cargo, as is the case for siRNA LNPs.5,7 This also means that while cryo-TEM can be used to determine the presence of siRNA, it is unable to quantify its interparticle distribution. The method typically provides two-dimensional projections of a three-dimensional structure, meaning that obtaining a complete, high resolution three-dimensional structure may be challenging. Some samples may also be difficult to image at near-atomic resolution, due to factors like sample heterogeneity and particle orientation distribution. Lastly, cryo-TEM requires skilled operators with expertise in sample preparation, data collection, and image analysis, and traditional cryo-TEM data analysis is known to be complex, computationally intensive, and time consuming. Fortunately, recent advances in automation are allowing the efficient acquisition and analysis of large datasets, supporting higher throughput cryo-TEM methodologies with fewer manual inputs.
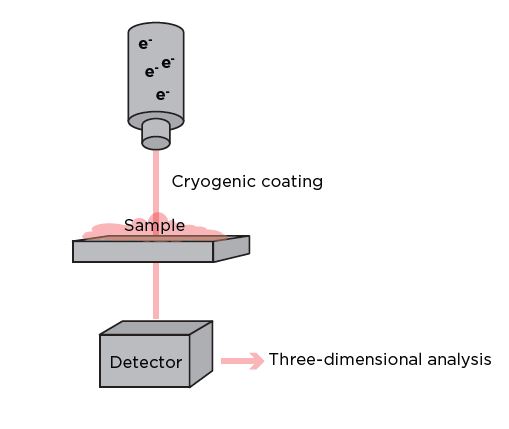
Figure 3: Cryo-TEM involves freezing a sample to retain its native structural information
3. Cylindrical illumination confocal spectroscopy (CICS)
CICS is a multi-laser, flow-based technique combining confocal microscopy with spectroscopy to obtain spatially resolved spectra from a sample. It incorporates a multi-color fluorescence spectroscopic technique with a single-molecule detection platform, fluorescence coincidence analysis, and a quantitative fluorescence deconvolution algorithm. CICS can be used to examine the mRNA and lipid contents in LNP formulations at the single-nanoparticle level. When passing the detection window, each LNP or free mRNA molecule in the solution generates a unique fluorescent burst signal, which is captured with single-fluorophore sensitivity. The raw data is then processed by a thresholding algorithm. Different species of interest can be determined by coincidence analysis of the fluorescent bursts, making it possible to quantify an LNP formulation’s mRNA payload capacity and distribution by deconvolving its signal distribution against that of the free mRNA. The fluorescence-based CICS technique is therefore able to differentiate free mRNAs, empty LNPs, and mRNA-loaded LNPs, as well as quantify the mRNA molecules encapsulated inside LNPs. It also sheds light on the effects of formulation parameters — such as the proportion of PEGylated lipids, the N:P ratio, the mRNA concentration, and mRNA size — on payload distribution and capacity.
General strengths of this analytical method include its improved spatial resolution compared to traditional microscopy — allowing for detailed analysis of LNP structure — and its optical sectioning capabilities, which enable researchers to selectively focus on specific planes within a three-dimensional sample. This reduces background signals and enhances the signal-to-noise ratio, leading to improved sensitivity in spectroscopic measurements for greater image clarity. On the other hand, confocal microscopy set-ups can be complex and may require skilled operators for optimal performance and data interpretation. Some confocal techniques may also require specific sample preparation, and samples need to be compatible with the chosen microscopy and spectroscopy modalities. The acquisition of confocal images and spectra can be relatively slow compared to some other methods, such as AUC, limiting overall throughput.
Figure 4: General set-up of a cylindrical illumination confocal spectroscope
4. Dynamic light scattering (DLS)
DLS provides information about the size and size distribution of particles, including the hydrodynamic radius. A laser beam is directed into the sample containing LNPs and, as the nanoparticles move due to Brownian motion, they cause fluctuations in the intensity of the scattered light. A detector measures these fluctuations over time, and the data obtained from the detector is analyzed using correlation functions. The autocorrelation function is particularly important in DLS analysis, as it provides information about the dynamic correlation length of the scattering particles, which is related to particle size. DLS software can then be used to analyze the decay of the autocorrelation function, extracting information about the distribution of particle sizes in the sample — the PDI.
Overall, DLS is a rapid and non-destructive characterization technique, and requires only a small sample volume, making it suitable for situations where the amount of LNP formulation is limited. It is also highly sensitive to changes in particle size, making it suitable for detecting variations in the PDI of LNPs. Additionally, DLS instruments are relatively easy to operate and the technique does not require extensive sample preparation, making it widely accessible to researchers with varying resources and experience levels.
The main downside of DLS for LNP characterization is its low resolution, which makes it unable to distinguish between empty and loaded LNPs of a similar size.5 This makes it less able to characterize loading heterogeneity than alternative analytical methods.5 DLS also provides an intensity-weighted size distribution, so larger particles contribute more to the signal, which can result in the masking of smaller particles. This means that it is most accurate and reliable for measuring more homogeneous samples, but performs less well for particles at the extremes of the size spectrum, such as very small nanoparticles or larger aggregates. Samples with a higher PDI, or those influenced by aggregate interference, may therefore require additional analysis with alternate techniques. DLS also provides information primarily about the hydrodynamic size of particles, offering limited insights into particle shape or morphology, and cannot differentiate between different components in a mixture. Consequently, it is often not possible to attribute the calculated PDI solely to LNPs in more complex formulations.
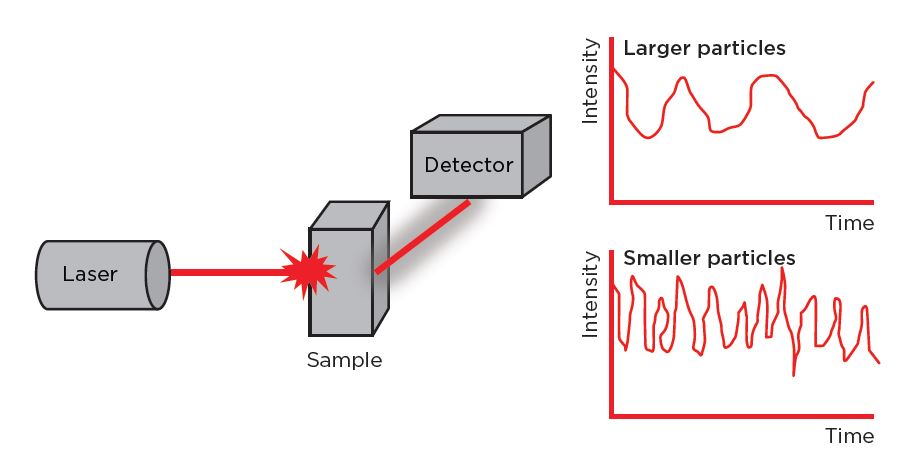
Figure 5. Schematic of particle size analysis by DLS. A laser illuminates particles undergoing Brownian motion in a sample. A photodetector measures the fluctuations in the intensity of scattered light over a time period. Stokes-Einstein’s equation is used to calculate the hydrodynamic diameter of the particles.
5. Microfluidic resistive pulse sensing (MRPS)
MRPS is based on the Coulter Principle,10 and is used as an orthogonal technique to flow cytometry in LNP characterization. MRPS is rapidly becoming more widely used in formulation optimization and in studying the impact of different parameters on numerous particle characteristics. The technology uses a microfluidic channel that contains an electrically insulating membrane with a small pore or constriction. An electric field is applied across this membrane, generating a measurable current, and particles passing through the constriction displace the electrolyte, causing temporary resistive pulses or drops in the electrical current, known as blockade events.11 The magnitude and duration of these pulses are proportional to the size and volume of the particle, and the frequency of pulses provides information regarding the concentration of particles in a sample.11 This data can in turn be used to calculate the size distribution, shape, surface charge and concentration of the individual LNPs in a formula.12
One major strength of the MRPS technique is its ability to provide high resolution direct measurements of multiple particle characteristics in real time as they pass through the microfluidic channel. This high resolution means that even subtle differences in particle size and payload can be determined, as well as the relative payload ratio, making the technique extremely precise. In addition, the accuracy of MRPS is independent of the refractive index, polydispersity and sample heterogeneity. Other key advantages of MRPS are its simplicity and ability to accurately analyze small sample volumes,12 which make the technique suitable for applications with limited sample availability.
On the other hand, the technique’s sensitivity is limited by the difficulties of fabricating a sufficiently small constriction, which is needed to provide high sensitivity. Small constrictions also limit the range of particle sizes that can be effectively measured using MRPS, and lead to slow sample transportation velocity through the microfluidic channel. This means that the throughput of portable MRPS devices is often relatively low.12 The applied electric field can also influence particle behavior, potentially leading to artifacts, and false signals may be generated according to the migration profile of the particles as they move across the microchannel.10 In addition, the MRPS method is more sensitive to changes in shape for non-spherical particles, potentially making it difficult to characterize LNPs with irregular morphologies.
A

B

C

Figure 6. A: Working principle of an MRPS system B: Individual particles generate signal as they each pass through nanoscale constriction C: Signal size correlates with the size of particle.
| ANALYTICAL METHOD | STRENGTHS | LIMITATIONS |
|---|---|---|
| AUC |
|
|
| Cryo-TEM |
|
|
| CICS |
|
|
| DLS |
|
|
| MRPS |
|
|
Table 1. A summary of the main advantages and disadvantages of five analytical techniques that can be used for LNP characterization.
Conclusion
The growing demand for novel LNP formulations is driving the need for high-throughput development and screening workflows. AUC is emerging as a strong method for LNP characterization. Beckman Coulter Life Sciences has pioneered this approach by introducing the first commercially available AUC sample characterization instrument and is committed to staying at the forefront of this dynamic field with its Optima AUC platform. This technology analyzes a wide array of particles in native, matrix-free conditions, and provides crucial data on parameters such as LNP morphology, mass, diameter, heterogeneity, mRNA loading ratio, and purity. All Optima systems have a high radial resolution and excellent signal-to-noise ratios, and enable the rapid and precise analysis of complex systems at discrete wavelengths for robust LNP formulation characterization.
References
- Cayman Chemical. A guide to lipid nanoparticle formulation: basic concepts & preparation procedures.
- Bepperling A, Richter G. Determination of mRNA copy number in degradable lipid nanoparticles via density contrast analytical ultracentrifugation. European Biophysics Journal. 2023;52(4-5):393-400. doi:10.1007/s00249-023-01663-y
- Sandhya M, Ramasamy D, Sudhakar K, Kadirgama K, Harun WSW. Ultrasonication an intensifying tool for preparation of stable nanofluids and study the time influence on distinct properties of graphene nanofluids – A systematic overview. Ultrason Sonochem. 2021;73:105479. doi:10.1016/j.ultsonch.2021.105479
- Bolze H, Riewe J, Bunjes H, Dietzel A, Burg TP. Continuous Production of Lipid Nanoparticles by Ultrasound-Assisted Microfluidic Antisolvent Precipitation. Chem Eng Technol. 2021;44(9):1641-1650. doi:10.1002/ceat.202100149
- Henrickson A, Kulkarni JA, Zaifman J, Gorbet GE, Cullis PR, Demeler B. Density Matching Multi-wavelength Analytical Ultracentrifugation to Measure Drug Loading of Lipid Nanoparticle Formulations. ACS Nano. 2021;15(3):5068-5076. doi:10.1021/ acsnano.0c10069
- Danaei M, Dehghankhold M, Ataei S, et al. Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems. Pharmaceutics. 2018;10(2):57. doi:10.3390/pharmaceutics10020057
- Li S, Hu Y, Li A, et al. Payload distribution and capacity of mRNA lipid nanoparticles. Nat Commun. 2022;13(1):5561. doi:10.1038/s41467- 022-33157-4
- Hald Albertsen C, Kulkarni JA, Witzigmann D, Lind M, Petersson K, Simonsen JB. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv Drug Deliv Rev. 2022;188:114416. doi:10.1016/j.addr.2022.114416
- NanoImaging Services. Seeing the Forest AND the Trees: Lipid Nanoparticle Analysis with Cryo-TEM . https://www. nanoimagingservices.com/blog/lnp-analyses-with-cryo-tem.
- Kim JS, Kwon SY, Lee JY, et al. High-throughput multi-gate microfluidic resistive pulse sensing for biological nanoparticle detection. Lab Chip. 2023;23(7):1945-1953. doi:10.1039/D2LC01064J
- Rozo AJ, Cox MH, Devitt A, Rothnie AJ, Goddard AD. Biophysical analysis of lipidic nanoparticles. Methods. 2020;180:45-55. doi:10.1016/J.YMETH.2020.05.001
- Song Y, Zhang J, Li D. Microfluidic and Nanofluidic Resistive Pulse Sensing: A Review. Micromachines (Basel). 2017;8(7):204. doi:10.3390/mi8070204



