Inactivation of COVID–19 Disease Virus SARS–CoV–2 with Beckman Coulter Viral RNA Extraction Lysis Buffers
Content Type: White PaperAuthor: Han Wei, Ph.D., Beckman Coulter Life Sciences. Indianapolis, IN
Background
The novel coronavirus 2019 (SARS–CoV–2), which was first reported in December 2019 in China, causes severe acute respiratory syndrome, including fever, cough, and difficulty breathing. It has been declared a pandemic by the World Health Organization due to its rapid spread in various countries and sufficient effect on human health1 . SARS–CoV–2, single–stranded RNA virus with nucleocapsid, belongs to the same family of viruses as SARS and MERS virus2. The nucleocapsid forms complexes with genomic RNA, and plays a critical role in enhancing the efficiency of virus transcription and assembly1.
Scientists around the world have developed methods for SARS–CoV–2 research detection. Currently, PCR–based techniques for the detection of SARS–CoV–2 viral RNA have been widely adopted in labs3. In June 2020, a sequencing–based SARS–CoV–2 research detection method was launched, thereby providing other research detection options. Working with highly infectious viruses such as SARS–CoV–2 are a threat to laboratory operators4, and thus, reliable inactivation of the virus from specimens is critical for lab safety.
RNA extraction is the essential first step for both PCR and sequencing–based methods. The recently launched RNAdvance Viral and RNAdvance Viral XP (Beckman Coulter Life Sciences) has demonstrated for SARS–CoV–2 RNA isolation from 200 µL of nasopharyngeal swab and saliva samples. To ensure the lab personnel can safely conduct RNA extraction using RNAdvance Viral or RNAdvance Viral XP, we conducted experiments to assess the virus inactivation status after treatment with lysis buffer (LBF) from RNAdvance Viral or RNAdvance Viral XP extraction kits.
Method
LBF Toxicity Test
To first determine the point at which LBF was no longer toxic to African green monkey kidney cells (Vero E6, ATCC CCL–81), LBF was first added to SARS–CoV–2 viruses and incubated at room temperature or 60oC for 20 minutes. After incubation, 0.1 mL of the virus–buffer mixture was diluted into 0.9 mL PBS, then 0.5 mL of the mixture was added to an Amicon filtration column. The columns were centrifuged at 10,000 RPM for 5 minutes. The flow–through (FT) was discarded, and the remaining sample was added to the column for another centrifugation at 10,000 RPM for 5 minutes. The FT was discarded, and 0.5 mL of PBS was added for buffer exchange and followed by centrifugation at 10,000 RPM for 5 minutes.
Each dilution was then added to confluent to Vero E6 cells (3 x 106) containing complete media (DMEM with 10% fetal bovine serum and antibiotics), incubated at 37°C with 5% CO2, and examined daily for 4 days. The lowest dilution at which LBF was no longer toxic (i.e., cells forming a visibly healthy monolayer similar to control wells or tubes) was considered as ground truth to avoid non–specific cytotoxicity in subsequent experiments.
SARS–CoV–2 Culture
SARS–CoV–2 (USA_WA1/2020 strain) was obtained from the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA, University of Texas Medical Branch). The WRCEVA virus aliquot was used to create SARS–CoV–2 stocks for inactivation studies treated with lysis buffer. All work with live virus was conducted in a biosafety level 3 (BSL3) laboratory.
To establish virus stocks, Vero E6 cells were infected one day post–plating at ~ 80% confluence using 1 mL of virus aliquot produced from initial infection to produce a “seed” stock. Cells were monitored under the microscope daily for evidence of virus–induced cytopathic effect (CPE) and a negative control without a virus was examined in parallel. When the culture showed ~ 70% CPE, the cells and culture media were transferred to 50 mL conical tubes. The cell debris was pelleted by centrifugation at 500 x g for 5 minutes. The supernatant was collected as the virus stock. An aliquot was tested by TCID50 assay and found to be 1.0 X 107 TCID50/mL. PCR analysis using the CDC assay showed the virus stock to be 3.9 X 109 genome copies (GC)/mL.
Inactivation of SARS–CoV–2 by Lysis Buffer
200 µL of SARS–CoV–2 viruses were mixed with 150 µL lysis buffer (LBF), and the samples were incubated at room temperature or 60°C for 20 minutes. After the incubation, 0.1 mL of the virus–buffer mixture was diluted into 0.9 mL PBS, then 0.5 mL of the mixture was added to an Amicon filtration column. The columns were centrifuged at 10,000 RPM for 5 minutes. The flow–through (FT) was discarded, and the remaining sample was added to the column for another centrifugation at 10,000 RPM for 5 minutes. The FT was discarded, and 0.5 mL of PBS was added for buffer exchange and followed by centrifugation at 10,000 RPM for 5 minutes. Finally, 0.2 mL of PBS was added to the column, and the sample was recovered for TCID50 assay. PBS was included in this experiment as a lysis buffer control.
Determination of TCID50
0.1 mL of sample was diluted into 0.9 mL of unsupplemented DMEM to generate the 10–fold dilution. A 10–fold serial dilution was made to achieve dilutions of 1 X 102 to 1 X 106. Subsequently, 0.1 mL of each dilution was added to 8 wells on a 96 well plate containing Vero E6 cells at 5,000 cells per well, which were plated the previous day in DMEM supplemented with 10% fetal bovine serum. Culture plates were incubated for 4 days at 37°C with 5% CO2. Plates were then examined under a microscope for the scoring of virus–induced CPE in each well. The plates were re–examined after 7 days, and data were recorded for each condition. TCID50/mL was calculated using the Reed and Muench method.
Infectivity of SARS–CoV–2 by Lysis Buffer
200 µL of SARS–CoV–2 viruses were mixed with 150 µL lysis buffer (LBF), and the samples were incubated at room temperature for 20 minutes. A day before inoculation, 3 x 106 Vero E6 cells were plated into a 6–well plate in 2 mL of complete culture media. On the day of the inoculation, 0.1 mL of the sample mixture was added to a single well of Vero E6 cells. The cultured cells were incubated at 37°C, with 5% CO2 for 5 days. After incubation, 0.5 mL of cell culture media was used to inoculate a well of 3 x 106 naïve Vero E6 cells in a 6–well plate. After an additional 5 days of incubation, cell morphology was observed to confirm evidence of either virus–induced CPE or buffer–associated toxicity. To determine whether the infectious virus could be detected in these samples, each sample was subjected to a TCID50 assay.
Results
LBF Toxicity
Despite the Amicon filtration of samples, the cytotoxicity was observed for the cells treated with 101 dilutions for LBF. However, no cytotoxicity was detected at 102 dilution and above. In order to avoid non–specific cytotoxicity in subsequent experiments, 101 dilutions for LBF was not included in the following experiments.
TCID50 Analysis
Measurable virus was detected for the 20 minutes at RT incubation conditions in the PBS negative control groups, but heating at 60°C for 20 minutes reduced the virus below the minimum measurable level for the assay (37 TCID50/mL). At either RT or 60°C treatment, no virus–induced CPE for the LBF conditions was observed and the calculated TCID50/mL is 316. There was a 2.3 Log reduction equal to a 215 fold viral activity reduction (TCID50/mL 794000/316). The results indicate that LBF from RNAdvance Viral or RNAdvance Viral XP reduces SARS–CoV–2 activity by at least 99.6%. Heat treatment (60°C) alone effectively reduces viral activity.
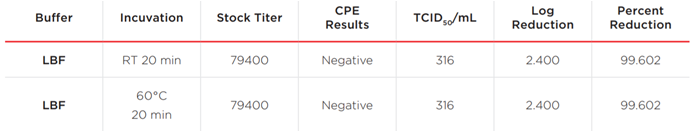
Table 1. Summary of viral titers, TCID50 and reduction of viruses tested.
Infectivity of SARS–CoV–2 by Lysis Buffer
No SARS–CoV–2 virus–induced CPE or cytotoxicity was observed. The result indicates that less than one infectious unit was present in the 0.1 mL inoculums of LBF–SARS–CoV–2 mixtures. A 3.90 Log reduction was calculated, which equals 99.987% reduction (1 (10/794000)).
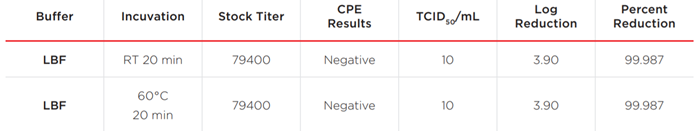
Table 2. Summary of infectivity of SARS–CoV–2 by lysis buffer.
Conclusion
This study demonstrates the ability of Lysis buffer (LBF) from RNAdvance Viral and RNAdvance Viral XP to inactivate the SARS–CoV–2 virus even at RT. The LBF toxicity data showed >99.6% effective viral inactivation. These results were consistent at both room temperature and 60°C for 20 minutes treated with LBF. The culture media TCID50 assay further improved the sensitivity to 99.987%. The assay sensitivity limits the confirmation of 100% inactivation due to the cytotoxic properties of LBF. The SARS CoV–2 virus is a novel virus, and no lysis buffer induced inactivation data is currently available. Taken together, this study provides significant information for laboratories when selecting nucleic acid extraction chemistries and conducting similar in–house inactivation experiments.
References
1. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS–CoV–2): An Overview of Viral Structure and Host Response. I Astuti, et al. Diabetes Metab Syndr. 2020.14(4): 407–412.
2. SARS–CoV–2, SARS–CoV, and MERS–COV: A Comparative Overview. AA Rabaan, et al. Infez Med. 2020.28(2):174–184.
3. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID–19 detection. Esbin MN, et al. RNA. 2020 26(7):771–783
4. High Contagiousness and Rapid Spread of Severe Acute Respiratory Syndrome Coronavirus 2. Sanche S, et al. Emerg Infect Dis. 2020. 26(7):1470–1477

