Automated library preparation for the MCI Advantage Cancer Panel at Miami Cancer Institute utilizing the Beckman Coulter Biomek i5 Span-8 NGS Workstation
Q Weiǂ, W Shenǂ, J Rantusǂ, A Medellin*, Z Smith*ǂMiami Cancer Center, Baptist Health South Florida, Florida International University, Miami, FL
*Beckman Coulter, Inc.
Introduction
Automated library preparation allows for higher throughput and less hands-on time for researchers and clinicians, particularly for complex next generation sequencing (NGS) assays. Here we describe the automation of the MCI Advantage Cancer Panel at Miami Cancer Institute, a 170 gene panel assay based upon the Illumina TruSight® Tumor 170 Panel, on the Beckman Coulter Biomek i5 Span-8 NGS Workstation.
Miami Cancer Institute of Baptist Health South Florida is a new state-of-the-art facility which brings outpatient cancer services together under one roof to offer world-class clinical services and cutting-edge therapies. Miami Cancer Institute is Florida's only member of the Memorial Sloan Kettering Cancer Alliance. The Molecular Diagnostics Laboratory is a clinical laboratory accredited by CLIA and CAP and provides cancer genomic and routine molecular testing, by means of complex molecular analyses in oncology and bone marrow transplantation. The laboratory serves the patients at Miami Cancer Institute as well as patients throughout Baptist Health South Florida. This laboratory supports multiple disciplines and also serves as a development hub for molecular testing, allowing the lab to serve Miami Cancer institute’s patients with cutting edge technology and expertise in routine as well as specialized esoteric molecular testing.

Figure 1: Illumina TruSight® Tumor 170 Manual Workflow1
The Illumina TruSight® Tumor 170 Panel covers 170 gene targets associated with solid tumors. The workflow includes the processing of both RNA and DNA libraries from a single sample over the course of two days (Figure 1). Following sequencing, single nucleotide variants (SNVs), indels, amplifications, gene fusions, and splice variants with a Mutant Allele Frequency as low as 5% can be identified with greater than 95% sensitivity and specificity1. The MCI Advantage Panel combines the Illumina TruSight® Tumor 170 Panel with bioinformatic analysis performed with the TST170 apps and Variant Interpreter on the HIPAA-compliant BaseSpace Enterprise platform in addition to orthogonal analysis utilizing the Philips IntelliSpace Genomics platform.

Figure 2: Miami Cancer Institute2
Illumina TruSight® Tumor 170 Panel Automated on the Biomek i5 Span-8
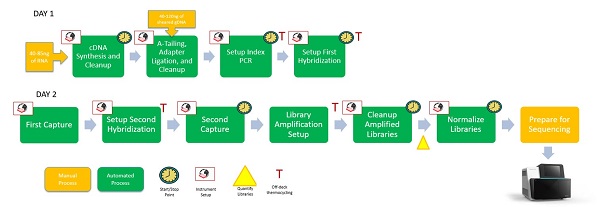
Figure 3: Automated TruSight® Tumor 170 workflow on the Biomek i5 Span-8 NGS Workstation
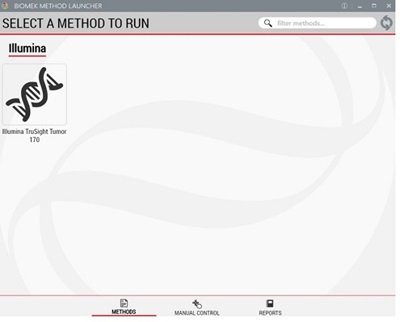
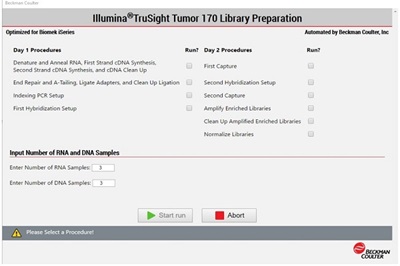
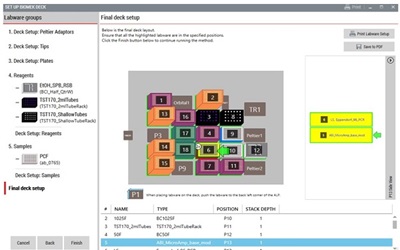
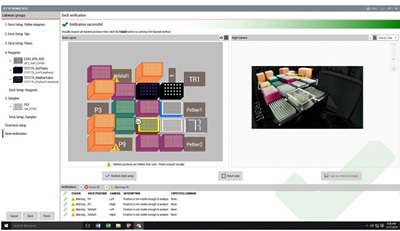
Figure 4: Demonstrated Method Interface consisting of Biomek Method Launcher dialog (A), Method Option Selector dialog (B), Guided Labware Setup dialog (C), and DeckOptix Final Check Verification (D).
As can be seen from Table 1, automation of the Illumina TruSight® Tumor 170 Panel on the Biomek i5 Span-8 liquid handler results in significant time savings and increased throughput compared to manual operators.
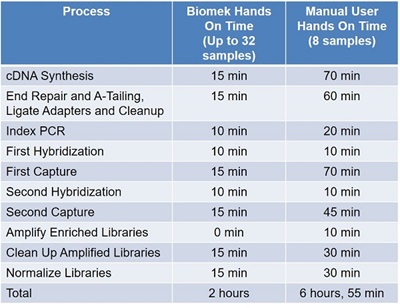
Table 1: Automated TruSight® Tumor 170 workflow on the Biomek i5 Span-8 NGS Workstation hands on time use compared to manual operator
Results
The automated Illumina TruSight® Tumor 170 method was installed and run at Miami Cancer Institute with a variety of samples ranging from Horizon Quantitative and Structural Reference Standards (Horizon, HD200 and HD789), Ashkenazim PGP Son Reference Standard (Horizon, GM24385), SeraSeq Fusion FFPE Reference (SeraCare, 0710–0129), and a number of in–house samples isolated with the FormaPure Total (Beckman Coulter). 108 DNA samples and 80 RNA samples were processed on the Biomek i5 Span–8 NGS Workstation over the course of 13 runs. Following sequencing, libraries were analyzed using the TruSight® Tumor 170 App on BaseSpace. Exon coverage at 100X or greater was very high across all DNA libraries (mean 99.78% with a standard deviation of 0.09%) as shown in Figure 5. RNA sequencing metrics (presented in Figures 6 and 7), show that for the 80 samples sequenced median insert length varied somewhat more than the DNA libraries (mean 109bp with a standard deviation of 14bp). RNA Median CV Coverage at 1000X was more consistent (mean 0.53 with a standard deviation of 0.09). RNA quality (utilizing the RNA Quality Number from the Advanced Analytical Fragment Analyzer) for the samples ranged from 10 (highest) to 1 (lowest) for the 59 samples that RQN values were available for. Purchased controls (samples 1–16) show a wide range of variability in terms of RQN as related to median insert length compared to the in–house samples (samples 17–65).
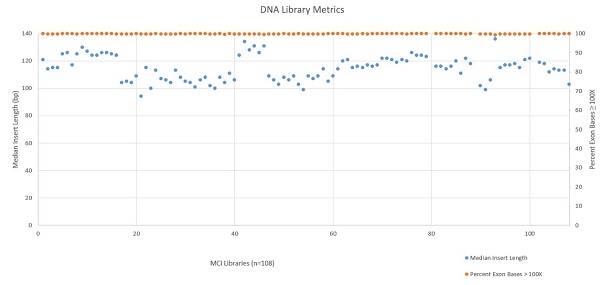
Figure 5: DNA Library Metrics
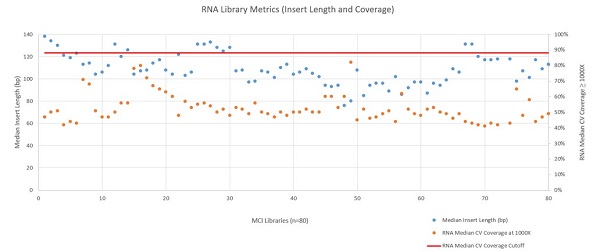
Figure 6: RNA Library Metrics (Insert Length and Coverage)
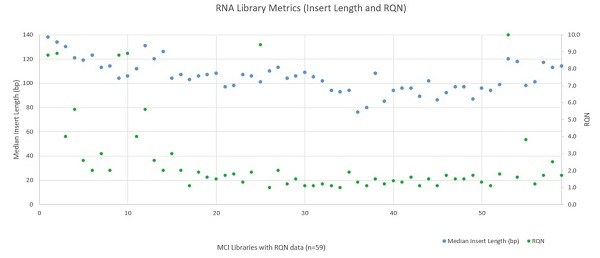
Figure 7: RNA Library Metrics (Insert Length and RQN)
83 of the samples processed using the MCI Advantage Cancer Panel were used for clinical validation. Testing using orthogonal methods such as High-Sensitivity Sanger Sequencing for EGRF, KRAS, and BRAF (LOD 10-15%) or FISH (ALK fusions) was performed at NeoGenomics. Concordance with MCI Advantage results was 98.7% Of the two discordant results one was determined to be concordant via RT-PCR while the second discordant result (an NRAS variant) was truly discordant.
Conclusion
In conclusion, we have shown that automation of the Illumina TruSight® Tumor 170 Panel on the Biomek i5 Span-8 NGS Workstation delivers libraries that yield quality results over a variety of sample inputs while saving valuable time compared to manual operators.
References
- https://support.illumina.com/content/dam/illumina-marketing/documents/products/datasheets/trusight-tumor-170-data-sheet-1170-2016-017.pdf
- Baptisthealth.net
Beckman Coulter makes no warranties of any kind whatsoever express or implied, with respect to this protocol, including but not limited to warranties of fitness for a particular purpose or merchantability or that the protocol is non-infringing. All warranties are expressly disclaimed. Your use of the method is solely at your own risk, without recourse to Beckman Coulter. Not intended or validated for use in the diagnosis of disease or other conditions. This protocol is for demonstration only, and is not validated by Beckman Coulter.

