A fully automated plate-based optimization of fed-batch culture conditions for monoclonal antibody-producing CHO cell line
Abstract
Immunoglobulin-based (IgG) therapies such as monoclonal antibodies (mAb) are increasingly used in the treatment of multiple human diseases. Process development for mAb entails laborious workflows that optimize medium and determine key cell culture conditions needed for increasing antibody titer production. Fed‑batch processes are applied in cell culture production to supply critical nutrients at physiological conditions. A combination of high‑performing basal medium with feed solution and the optimal feed regimen are often combined for a given cell line to boost high-antibody titers. Given the unique combinations needed to establish culture conditions, design of experiments (DoE) can be used to statistically determine the optimal fed-batch combinations based on randomization. DoE-based workflows produce statistically relevant high throughput analysis that can be time consuming and error prone.
Here we describe an automated and miniaturized 96-well plate-based system for suspension cell culture designed to match the high demand for high-throughput applications in biopharmaceutical cell-based process development workflows for therapeutic development. The workflow applies unique combinations of batch feeds generated using a full factorial (2n) DoE method that can be evaluated across various basal cell culture media for IgG-producing CHO cell line. Next, an automated workflow was designed for media optimization experiments that reduces hands-on time yet increases walk-away time, decreases the total time needed for media optimization, and performs hands-free fed-batch screening, viable cell, and IgG quantifications.
An IgG-producing CHO cell line was cultured in Forti-CHO medium, and individual HyClone Cell Boost™ feed supplements were evaluated in spiked batch and fed-batch experiments. Basal medium was spiked with feed combinations using Cell Boost HyClone reagents starting on day 0 of batch cultures, followed by frequent analysis for cell viability and IgG quantity for 240 hours. Candidate Cell Boost supplements were thereafter applied in fed-batch cultures establishing their relative ratios. Various fed-batch culture conditions supported variable mAb titers that were consistent with media optimization and fed-batch screening. This workflow provides opportunities for automated high-throughput analysis of DoE-based screening of fed-batch conditions for process development of monoclonal antibody production in a hands-free environment.
Introduction
Immunoglobulin-based (IgG) therapies such as monoclonal antibodies (mAb) are increasingly used as therapeutics for treatment of various human diseases. There is also an increase in demand for advanced cell culture process automation that can enhance throughput capacity and improve efficiency of process development that yields high antibody titers during mAb expansion. Process development for mAb entails laborious workflows that optimize medium and determine key cell culture conditions needed for producing high-antibody titers.1 Fed‑batch processes are applied in cell culture production to supply critical nutrients at physiological conditions. A combination of high‑performing basal medium with feed solution and the optimal feed regimen are often combined for a given cell line to boost high antibody titers.1 To accelerate process development and improve efficiency of optimization, streamlined and highly parallel workflows are needed.
Process development for enhancing IgG titer production can be accomplished by a variety of approaches including screening reagents, media optimizations and fed-batch analysis. A combination of a variety of cell culture reagents and media can play key roles in the determination of the unique cell culture conditions that enhance levels of antibody titers. Applying a rigorous method such as design of experiments (DoE) can statistically and reliably determine the optimal fed-batch combinations through randomized groupings of candidate factors for experimental validation. However, depending on the number of factors needed for validation, the DoE generated combinations can also yield multiple variables whose experimental validation can be labor intensive and extremely costly. This is especially true given that mammalian cell lines are typically used as technological vehicle production that require large volumes of media when grown in cell culture flasks. For instance, the routinely used shaker flasks that require large volumes of media (10 mL to 3 L), are resource intensive and can limit the total number of experimental variables, thus reducing the design window and potentially eliminating useful factors that enhance product development.
An automated and miniaturized 96-well plate-based system for suspension cell culture was therefore designed to match the high demand for high-throughput applications in biopharmaceutical process development that accommodates DoE-based fed-batch screening and media optimizations. Combinations of batch feeds were generated using full factorial (2n) DoE and evaluated across various basal cell culture media for IgG-producing CHO cell line. An automated workflow was designed for media optimization experiments that minimize hands-on time to minimize unintended human errors yet increase walk-away time while decreasing the total time needed for product development.
Thus, this application note demonstrates an applicable and efficient automated workflow for establishing optimal feed combination for best performing fed-batch processes using a 96-deepwell plate. The workflow contains the use of an integrated liquid handler – a Biomek i7 hybrid workstation – an automated cell counter – the Vi-CELL BLU cell viability analyzer – a SpectraMax i3 plate reader, and a Kuhner shaker incubator (Figure 1). These systems allow for automation of an entire media optimization workflow, including transfer of cells and cell culture medium, fed-batch screening, and feed combinations, automated viable cell quantification, and quantification of IgG production from the viable cells using the Valita Titer system. Moreover, automated data tracing is accomplished using the Biomek data acquisition and reporting tool (DART), which gathers data and synthesizes runtime information from Biomek log files, capturing each manipulation of the sample across integrated systems during the Biomek method run.

Figure 1: Key technology for development of microplate systems for cell-based process development such as media optimization for IgG development with scale-up potential.
Methods
Automated cell culture conditions for full factorial DoE media optimization
The Chinese Hamster Ovary cell Agarabi CHO cell line (ATCC cat# CRL-3440) engineered to express human IgG1 and adapted to suspension culture were grown in Gibco™CD FortiCHO (ThermoFisher Scientific cat# A1148301) basal medium containing 1% anti-clamping reagent and 200 nM glutamax at 37°C Kuhner incubator and 8% CO2. For process development validation, Cell Boost reagents from Cytiva were evaluated using predesigned DoE matrix (Figure 2A). The DoE matrix was generated using JMPR software using a full factorial approach to validate 5 factors (Cell Boost 1, 2, 3, 7A and 7B) in cell culture medium (Figure 2A). Using full factorial DoE with 2n factors, where n=5, 32 random variables were generated and designed to be evaluated using 3 independent biological triplicates (Figure 2A-B). Cell Boost media were combined in a 1:1 volume-by-volume combination for a total volume of 90 μL Cell Boost reagents. Cells were added to a total density of 0.5x106 cells/mL and topped with basal media for a total cell culture volume of 700 μL per run. Cells were cultured in sterile 96-well deepwell plates (Beckman Coulter Life Sciences, product #267007) covered with 96-deepwell sandwich covers (Kuhner cat# SMCR1296c), that contain 0.8 mm holes that allow for oxygen distribution throughout the plate during cell culture incubation. Cells were incubated in Kuhner incubator at 37°C, 8% CO2, 50 mm orbital diameter and a shaking speed of 225 RPM (Figure 1). Cell viability was quantified using a Vi-CELL BLU automated cell counter (Beckman Coulter Life Sciences product #C19196) designed to accommodate 96-well sample plates (Figure 1). Cells were collected at various time points, and analysis of both viable cell density and IgG quantification was performed. Data were transferred into DoE JMPR software for downstream analysis and modelling evaluation.
Automated cell seeding
Automated workflow was designed using Biomek software. Key automation processes included cell seeding at timepoint 0 hours (T0), transfer of Cell Boost reagents (Figure 2B), collection of volume for quantification of viable cells and automated analysis of IgG levels using the Valita Titer kit. During the Biomek method design, Data Acquisition and Reporting Tool (DART) was configured to log all method details, errors, pipetting and transfers (Figure 3A, red arrow). This configuration enables Biomek software to log all corresponding log files, which can be viewed and automatically analyzed in DART report builder without any copy-paste of run details across all integrated systems.

Figure 2: Full factorial DoE matrix to evaluate effects of 5 different Cell Boost reagents on IgG production in CHO cells.
Automated IgG analysis by Valita Titer Assay
Automation of Valita Titer assay was performed following assay details provided in the manufacturer instructions and as previously described.2 The liquid handling steps were executed with an automated method on a Biomeki7 Hybrid workstation (Figure 3A). First, 60 μL of assay buffer (Forti-CHO) was added to each well of the black, 96-well, Valita Titer assay plate, followed by a 5-minute incubation step with shaking using the on-deck Orbital Shaking ALP. Standard curves were generated by serially diluting human IgG antibody from 100 mg/L to 1 mg/L in Forti-CHO buffer. Next, 60 μL of the standard curve samples was added to the Valita Titer assay plate, and the plate was shaken for 5 minutes. Finally, the plate was transferred to an integrated SpectraMax i3x (Molecular Devices) equipped with a fluorescence polarization detection cartridge, the plate was incubated in the dark for 3 minutes, and polarization was measured. Analysis of unknown samples was performed by the same approach, where media was harvested from cell culture samples by centrifugation followed by automated Valita Titer assay analysis. Sample harvesting was performed at timepoints 0, 96 and 144 hours for downstream analysis.
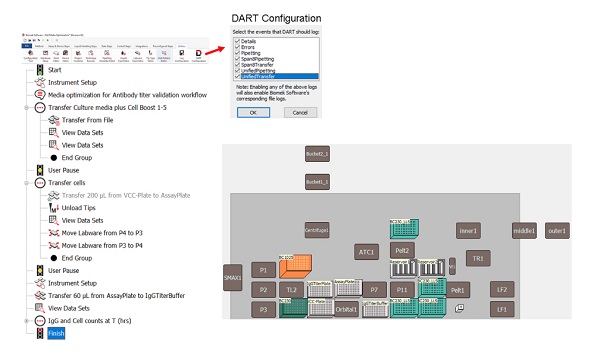
Figure 3: Automated Biomek method and deck layout for hands-free cell seeding, batch-feds, and IgG quantification.
Results and discussion
Cell viability in Cell Boost spiked samples
The key objective here was to evaluate the automation of media optimization workflows for process development used for enhancing IgG titer production. Thus, a DoE approach was initially used to determine combination strategy of fed-batch reagents needed for evaluation of IgG titer production (Figure 2A-B). An automated method was designed for performing cell seeding, spiking of fed-batch reagents and quantification of viable cell counts by fed-batch treatment strategy (Figure 2B, 3A-B). Viability of cells across different fed-batch spiking was analyzed at various time points ranging from (0-200 hours) post Cell Boost spiking. At 0 hours, 6 different 96-deepwell plates were each seeded with 0.5x106 cells/mL and spiked with Cell Boost reagents as per DoE design (Figure 2A-B). Cell incubation was initiated at the same time and a plate was isolated every 24-48 hours to quantify cell viability as per the pre-defined schematic (Figure 4A). The data generated demonstrated that although cells were incubated in different plates and analyzed at different time points, there was consistent reproducibility of cell proliferation rates as per the type of media and spiked Cell Boost reagents. The growth curves for all spiked factors demonstrated variability in cell proliferation rates by treatment and basal media conditions (Figure 4B). To understand these effects in detail, data analysis was performed by grouping the experimental factors together (Figure 4C-F). By this analysis we found that groups containing addition of a single factor did not produce significant differences in proliferation rates compared to the basal medium (Figure 4C). One separation was observed in the cohort containing two Cell Boost treatments, where cells treated with Cell Boost 3 and 7B had decreased proliferation rates compared to the other 2-group factors and basal medium. Three or more Cell Boost treatments retained cell viability at levels like baseline with various conditions leading to increased proliferation (Figure 4E-F) and two conditions leading to decreased proliferation (Figure 4F).
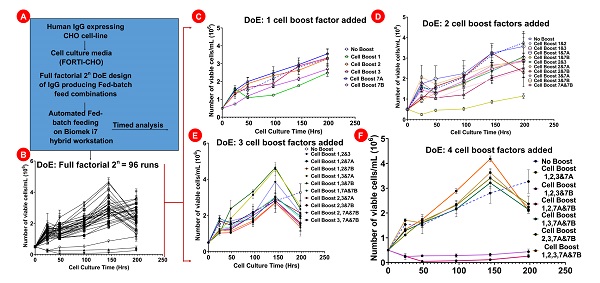
Figure 4: Quantification of viable cells. A) Workflow schematic. B) Cell proliferation across all 32 samples performed in triplicate for a total of 96 samples. C) Comparison of proliferation between baseline medium versus medium spiked with a single Cell Boost reagent. D) Proliferation between baseline medium versus medium spiked with two Cell Boost reagents. E) Proliferation between baseline medium versus medium spiked with three Cell Boost reagents. F) Proliferation between baseline medium versus medium spiked with four Cell Boost reagents.
These vital observations suggested that plate-based cell seeding can be performed using Biomek liquid handler and this can be performed using both a multichannel and Span-8 pods as performed in the current workflow. Moreover, cell viability can be performed by removing plates from incubation, using automated liquid transfer to a cell counter plate and re-incubating, or re-seeding the plate for additional biological manipulation as needed. The Biomek liquid handling steps did not affect the health of the cells, suggesting that an automated workflow could play a major role in meeting high-throughput demands and validation of unique DoE matrices for multi-variable analysis for cell-based assays.
Validation of automated Valita Titer workflow
Automation of Valita Titer assay was performed following assay details provided in the manufacturer instructions. To evaluate the accuracy of the automated Valita Titer method, standard curves were generated using the Biomek i7 hybrid workstation (Figure 5). The control human IgG was tested at concentrations ranging from 100 mg/L to 1 mg/L in triplicate, followed by measurement of fluorescence polarization. This yielded an R2 value of 0.9919 (Figure 5).

Figure 5: Standard Curve Analysis.
Cell Boost treatment effects on IgG titer
The automated IgG quantification workflow was then designed to perform an IgG standard curve for each plate containing samples with unknown levels of IgG from various cell culture conditions as per the DoE format and Cell Boost spiking. For each plate, a standard curve was generated to estimate IgG levels from unknown samples. The data generated was directly imported into the DoE software to generate models and analyzed effects of Cell Boost treatment on IgG titer production, cell viability and to relate cell viability to IgG titer levels. This led to a unique predictive model that closely estimates the amount of IgG titer production for each Cell Boost reagent (Figure 6A).
Based on this model, it is suggested that Cell Boost reagents 7a/b uniquely enhance IgG levels when combined with any of the Cell Boost reagents 1-3 and spiked into Forti-CHO medium containing Agarabi-CHO monoclonal antibody producing cell line (Figure 6A). Moreover, evaluation of cell viability by Cell Boost treatment suggested that Cell Boost 3, 7a and 7b yielded the highest viable cell density that also contributed to a high amount of IgG titer production (Figure 6B). These vital observations suggest that an automated cell culture system containing automation technologies such as those in Figure 1 can play a significant role in increasing the throughput capacity of cell-based workflows designed for IgG production. Importantly, these technologies provide capacity for assay miniaturization and can contribute to lower cost of product development, reagent validation and media optimization necessary to screen and develop therapeutic antibodies.

Figure 6: Predictive DoE model for media optimization and IgG production by automated workflow. A) Output of full factorial DoE combinations of Cell Boost reagents and model of predicting IgG production by Cell Boost combination or single reagent spikes. Inset shows statistical analysis of each Cell Boost reagent and probability of enhancing IgG titer levels. B) Analysis of each Cell Boost effect on cell viability and maximum IgG titer levels.
Automated data tracing by data acquisition and reporting tool (DART)
Performing automated cell-based assays in which multiple technologies are integrated to collectively validate different components of the workflow can lead to excessive amounts of data generation that require careful management, storage and retrieval. In the current DoE workflow, data tracing was performed using a data management software - data acquisition and reporting tool (DART) – that is designed to work with Biomek software and ensure data continuity across systems that are integrated with a Biomek workstation. Therefore, an integrated system with DART software can access a DART repository, a database designed to store details of Biomek methods and runs. DART stores method details for both successful and unsuccessful runs, pinpointing key details such as dates, times, deck positions, plates, volumes, plate barcodes, as well as movement of plates from Biomek deck to and from integrated analyzers/ instruments (Figure 7). DART report builder function provides automated data tracing that minimizes error prone copy-and-paste approach, and allows storage details to be available in a database that can be accessed remotely and directly transferred into Microsoft Excel.

Figure 7: Automated data tracing using DART software. A view of the DART database for the automated DoE media optimization workflow tracing data from the method runs, labware details, well details and reports.
The DART software also allows storage of each method run and number of times the method has been accessed, providing key details including run times, detected errors, and transfer details, among others, for both successful and unsuccessful runs (Figure 7). Comparisons of related methods, or comparisons of successful versus unsuccessful runs, can be performed within the DART data browser, which can trace essential information regarding each method run and compare each stage of the method and whether revisions have been made from run-to-run (Figure 8). This can guide in tracing any changes that may have resulted in different observations throughout the history of the data collection for a given Biomek method (Figure 8).
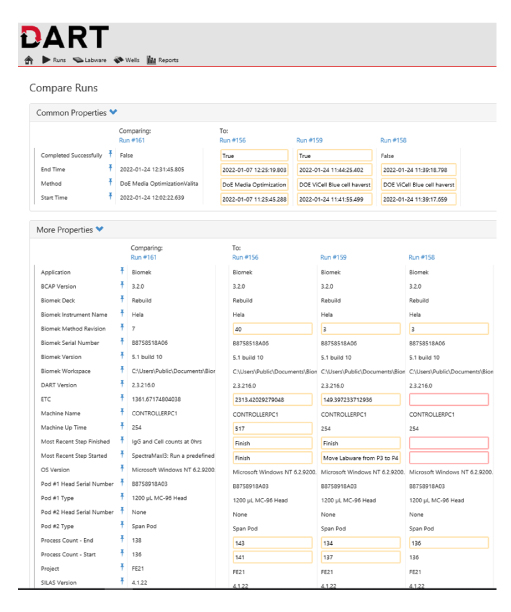
Figure 8: Example method run comparison with the DART software. A view of the DART method run comparisons for the automated DoE media optimization workflows for two successful and two unsuccessful runs.
Summary
In sum, this application note describes an automated workflow encompassing various Biomek integrated systems, together with cell culture equipment and technology needed to perform hands-free cell-based methods such as media optimization during production of monoclonal antibody titers. The workflow utilizes full factorial design of experiment approach to characterize best performing fed-batch reagents and media combinations that enhance production of high IgG titers, generating a predictive DoE-based model that culminates in statistical decision making and determination of unique Cell Boost reagents that guide cell-specific titer production in a monoclonal IgG producing CHO cell line. In addition to performing the workflow in an automated setting, we demonstrate the application of automated data tracing software, DART, which is configured to work with the Biomek software collecting key Biomek method run details and automatically deposit them in a remotely accessible DART repository database for storage and easy access. A combination of networked Biomek hybrid workstation systems with DART data tracing capabilities thus provides automation of both liquid handling steps as well as data handling and storage steps for Biomek systems and integrated systems needed for cell-based drug discovery workflows.
References
1. Markert S, Joeris K. Establishment of a fully automated microtiter plate-based system for suspension cell culture and its application for enhanced process optimization. Biotechnology Bioengineering. 2017 114(1):113-121. doi: 10.1002/bit.26044. PMID: 27399304.
2. Michael Hayes, Automation of IgG Quantification using the Biomek i7 Hybrid Automated Workstation. www.beckman.com content code 21.09.2634.AUTO.
Equipment and Material
| Equipment | Manufacturer |
|---|---|
| Biomek i7 hybrid automated liquid handler | Beckman Coulter Life Sciences |
| SpectraMax i3x plate reader | Molecular Devices |
| Orbital shaker | Benchmark Scientific |
| 37°C Kuhner shaker incubator 7 | Kuhner Shaker Inc |
| Vi-CELL BLU cell viability analyzer | Beckman Coulter Life Sciences |
| Reagents | Manufacturer | Part Number |
|---|---|---|
| Valita Titer Assay | Beckman Coulter Life Sciences | VAL003 |
| CD-Fort-CHO | ThermoFisher Scientific | A1148301 |
| Cell Boost 1 | Cytiva | SH31113.02 |
| Cell Boost 2 | SH31114.02 | |
| Cell Boost 3 | SH31115.01 | |
| Cell Boost 7A | SH31119.01 | |
| Cell Boost 7B | SH31120.03 |
| Consumables | Manufacturer | Part Number |
|---|---|---|
| BC230 Pipette Tips | Beckman Coulter Life Sciences | B85906 |
| BC1070 Pipette Tips | B85945 | |
| BC1025 Pipette Tips | B85981 | |
| VI-CELL BLU reagent kit | C06019 |

